3A66
 
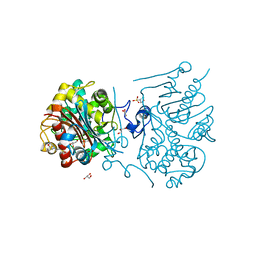 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/H266N/D370Y mutant with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOATE-DIMER HYDROLASE, 6-AMINOHEXANOIC ACID, ... | | Authors: | Kawashima, Y, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2009-08-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzymatic Synthesis of Nylon-6 Units in Organic Sol Contained Low-Water: Structural Requirement of 6-Aminohexanoate-Dimer Hydrolase for Efficient Amid Synthesis
To be Published
|
|
1BFG
 
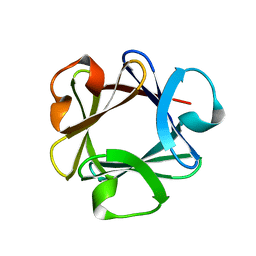 | | CRYSTAL STRUCTURE OF BASIC FIBROBLAST GROWTH FACTOR AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Kitagawa, Y, Ago, H, Katsube, Y, Fujishima, A, Matsuura, Y. | | Deposit date: | 1993-04-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of basic fibroblast growth factor at 1.6 A resolution.
J.Biochem.(Tokyo), 110, 1991
|
|
3WKR
 
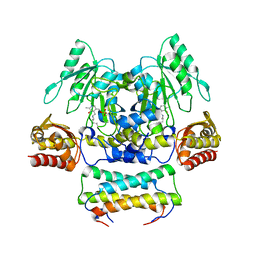 | | Crystal structure of the SepCysS-SepCysE complex from Methanocaldococcus jannaschii | | Descriptor: | O-phospho-L-seryl-tRNA:Cys-tRNA synthase, Uncharacterized protein MJ1481 | | Authors: | Nakazawa, Y, Asano, N, Nakamura, A, Yao, M. | | Deposit date: | 2013-10-30 | | Release date: | 2014-07-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Ancient translation factor is essential for tRNA-dependent cysteine biosynthesis in methanogenic archaea.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WKS
 
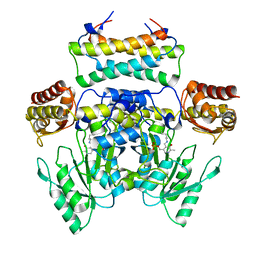 | | Crystal structure of the SepCysS-SepCysE N-terminal domain complex from | | Descriptor: | O-phospho-L-seryl-tRNA:Cys-tRNA synthase, Uncharacterized protein MJ1481 | | Authors: | Nakazawa, Y, Asano, N, Nakamura, A, Yao, M. | | Deposit date: | 2013-10-30 | | Release date: | 2014-07-30 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (3.029 Å) | | Cite: | Ancient translation factor is essential for tRNA-dependent cysteine biosynthesis in methanogenic archaea.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3VMH
 
 | | Oxygen-bound complex between oxygenase and ferredoxin in carbazole 1,9a-dioxygenase | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin component of carbazole, ... | | Authors: | Ashikawa, Y, Nojiri, H. | | Deposit date: | 2011-12-12 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insight into the substrate- and dioxygenbinding manner in the catalytic cycle of rieske nonheme iron oxygenase system, carbazole 1,9adioxygenase
Bmc Struct.Biol., 12, 2012
|
|
3VMG
 
 | | Reduced carbazole-bound complex between oxygenase and ferredoxin in carbazole 1,9a-dioxygenase | | Descriptor: | 9H-CARBAZOLE, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ashikawa, Y, Nojiri, H. | | Deposit date: | 2011-12-12 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insight into the substrate- and dioxygenbinding manner in the catalytic cycle of rieske nonheme iron oxygenase system, carbazole 1,9adioxygenase
Bmc Struct.Biol., 12, 2012
|
|
7BON
 
 | | Crystal structure of recombinant horse spleen apo-R52C/E56C/R59C/E63C-Fr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Maity, B, Ito, N, Abe, S, Lu, D, Ueno, T. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Design of Multinuclear Gold Binding Site at the Two-fold Symmetric Interface of the Ferritin Cage
Chem Lett., 49, 2021
|
|
7BOM
 
 | | Crystal structure of recombinant horse spleen apo-R52C/E56C/R59C/E63C-Fr immobilized with gold ions. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Maity, B, Ito, N, Abe, S, Lu, D, Ueno, T. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design of Multinuclear Gold Binding Site at the Two-fold Symmetric Interface of the Ferritin Cage
Chem Lett., 49, 2021
|
|
3VMI
 
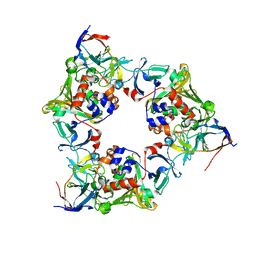 | | Carbazole- and oxygen-bound complex between oxygenase and ferredoxin in carbazole 1,9a-dioxygenase | | Descriptor: | 9H-CARBAZOLE, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ashikawa, Y, Nojiri, H. | | Deposit date: | 2011-12-12 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into the substrate- and dioxygenbinding manner in the catalytic cycle of rieske nonheme iron oxygenase system, carbazole 1,9adioxygenase
Bmc Struct.Biol., 12, 2012
|
|
3VSM
 
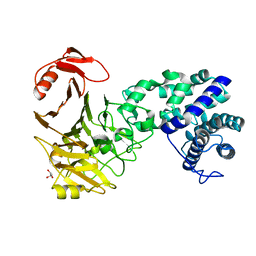 | | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein | | Descriptor: | GLYCEROL, Occlusion-derived virus envelope protein E66 | | Authors: | Kawaguchi, Y, Sugiura, N, Kimata, K, Kimura, M, Kakuta, Y. | | Deposit date: | 2012-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein
To be Published
|
|
3VSN
 
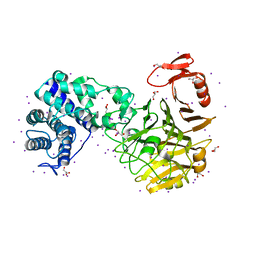 | | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein | | Descriptor: | GLYCEROL, IODIDE ION, Occlusion-derived virus envelope protein E66 | | Authors: | Kawaguchi, Y, Sugiura, N, Kimata, K, Kimura, M, Kakuta, Y. | | Deposit date: | 2012-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein
To be Published
|
|
3WUQ
 
 | | Structure of the entire stalk region of the dynein motor domain | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1 | | Authors: | Nishikawa, Y, Oyama, T, Kamiya, N, Kon, T, Toyoshima, Y.Y, Nakamura, H, Kurisu, G. | | Deposit date: | 2014-05-01 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the entire stalk region of the Dynein motor domain
J.Mol.Biol., 426, 2014
|
|
3VWL
 
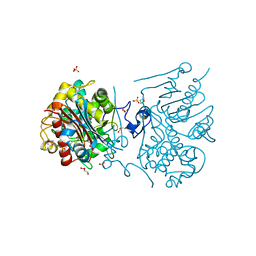 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187S/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWR
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187G/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWM
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187A/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWQ
 
 | | 6-aminohexanoate-dimer hydrolase S112A/G181D/R187A/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
3VWP
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187S/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
3VWN
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187G/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
2WOQ
 
 | | Porphobilinogen Synthase (HemB) in Complex with 5-acetamido-4- oxohexanoic acid (Alaremycin 2) | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ALAREMYCIN 2, ... | | Authors: | Heinemann, I.U, Schulz, C, Schubert, W.-D, Heinz, D.W, Wang, Y.-G, Kobayashi, Y, Awa, Y, Wachi, M, Jahn, D, Jahn, M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the heme biosynthetic Pseudomonas aeruginosa porphobilinogen synthase in complex with the antibiotic alaremycin.
Antimicrob. Agents Chemother., 54, 2010
|
|
2YRN
 
 | | Solution structure of the CH domain from Human Neuron navigator 2 | | Descriptor: | Neuron navigator 2 isoform 4 | | Authors: | Tomizawa, T, Tochio, N, Koshiba, S, Inoue, M, Nakamura, Y, Furukawa, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain from Human Neuron navigator 2
To be Published
|
|
2RPS
 
 | | Solution structure of a novel insect chemokine isolated from integument | | Descriptor: | Chemokine | | Authors: | Kamiya, M, Nakatogawa, S, Oda, Y, Kamijima, T, Aizawa, T, Demura, M, Hayakawa, Y, Kawano, K. | | Deposit date: | 2008-07-28 | | Release date: | 2009-06-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A novel peptide mediates aggregation and migration of hemocytes from an insect
Curr.Biol., 19, 2009
|
|
5ZPD
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 288 K (1) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPC
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 283 K (4) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.741 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZP7
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 277 K (3) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8H1P
 
 | | Cryo-EM structure of the human RAD52 protein | | Descriptor: | DNA repair protein RAD52 homolog | | Authors: | Kinoshita, C, Takizawa, Y, Saotome, M, Ogino, S, Kurumizaka, H, Kagawa, W. | | Deposit date: | 2022-10-03 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | The cryo-EM structure of full-length RAD52 protein contains an undecameric ring.
Febs Open Bio, 13, 2023
|
|
