3R2K
 
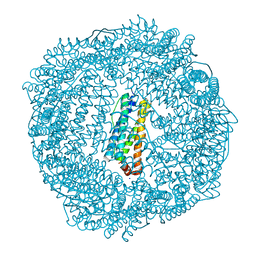 | | 1.55A resolution structure of As-Isolated FtnA from Pseudomonas aeruginosa (pH 7.5) | | Descriptor: | Bacterioferritin, SODIUM ION | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
4ILO
 
 | | 2.12A resolution structure of CT398 from Chlamydia trachomatis | | Descriptor: | 1,2-ETHANEDIOL, CT398, ZINC ION | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2012-12-31 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | 2.12A resolution structure of CT398 from Chlamydia trachomatis
To be Published
|
|
3D7R
 
 | | Crystal structure of a putative esterase from Staphylococcus aureus | | Descriptor: | BROMIDE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lam, R, Battaile, K.P, Chan, T, Romanov, V, Lam, K, Soloveychik, M, Wu-Brown, J, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2008-05-21 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of a putative esterase from Staphylococcus aureus
To be Published
|
|
4JBU
 
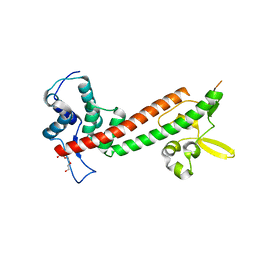 | | 1.65A structure of the T3SS tip protein LcrV (G28-D322, C273S) from Yersinia pestis | | Descriptor: | TETRAETHYLENE GLYCOL, Virulence-associated V antigen | | Authors: | Lovell, S, Chaudhury, S, Battaile, K.P, Plano, G, De Guzman, R.N. | | Deposit date: | 2013-02-20 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the Yersinia pestis tip protein LcrV refined to 1.65A resolution
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4JPO
 
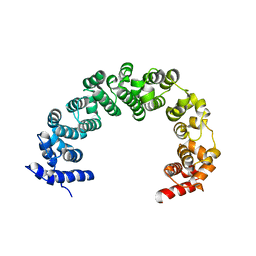 | | 5A resolution structure of Proteasome Assembly Chaperone Hsm3 in complex with a C-terminal fragment of Rpt1 | | Descriptor: | 26S protease regulatory subunit 7 homolog, DNA mismatch repair protein HSM3 | | Authors: | Lovell, S, Battaile, K.P, Singh, R, Roelofs, J. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Reconfiguration of the proteasome during chaperone-mediated assembly.
Nature, 497, 2013
|
|
4JER
 
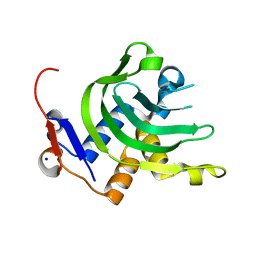 | | 1.1A resolution Apo structure of the hemophore HasA from Yersinia pestis (Tetragonal Form) | | Descriptor: | Hemophore HasA, SODIUM ION | | Authors: | Kumar, R, Lovell, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Hemophore HasA from Yersinia pestis (HasAyp) Coordinates Hemin with a Single Residue, Tyr75, and with Minimal Conformational Change.
Biochemistry, 52, 2013
|
|
4ILQ
 
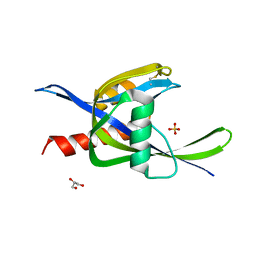 | | 2.60A resolution structure of CT771 from Chlamydia trachomatis | | Descriptor: | CT771, GLYCEROL, SULFATE ION | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2012-12-31 | | Release date: | 2014-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chlamydia trachomatis CT771 (nudH) Is an Asymmetric Ap4A Hydrolase.
Biochemistry, 53, 2014
|
|
3R2R
 
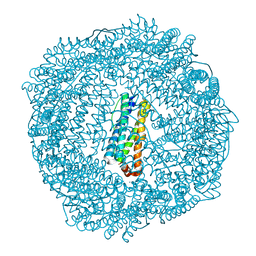 | | 1.65A resolution structure of Iron Soaked FtnA from Pseudomonas aeruginosa (pH 6.0) | | Descriptor: | Bacterioferritin, FE (III) ION, SODIUM ION, ... | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
3R2M
 
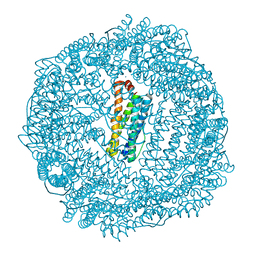 | | 1.8A resolution structure of Doubly Soaked FtnA from Pseudomonas aeruginosa (pH 7.5) | | Descriptor: | Bacterioferritin, FE (III) ION, SODIUM ION | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
3Q7S
 
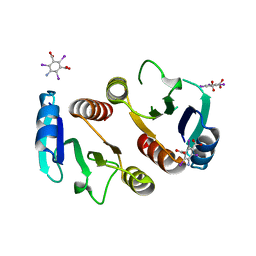 | | 2.1A resolution structure of the ChxR receiver domain containing I3C from Chlamydia trachomatis | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Transcriptional regulatory protein | | Authors: | Hickey, J, Lovell, S, Battaile, K.P, Hu, L, Middaugh, C.R, Hefty, P.S. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The atypical response regulator protein ChxR has structural characteristics and dimer interface interactions that are unique within the OmpR/PhoB subfamily.
J.Biol.Chem., 286, 2011
|
|
7TQ7
 
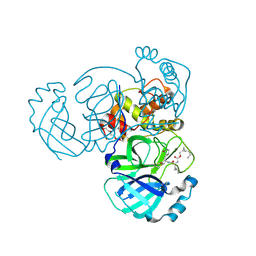 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 13c | | Descriptor: | N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucinamide, Orf1a protein, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ6
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 13d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ5
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 10d | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ8
 
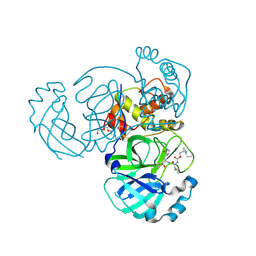 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 14d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, Orf1a protein, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ2
 
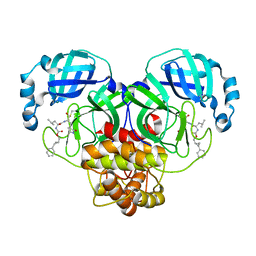 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 1c | | Descriptor: | 3C-like proteinase, N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-phenylcyclopropyl]methoxy}carbonyl)-L-leucinamide | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ3
 
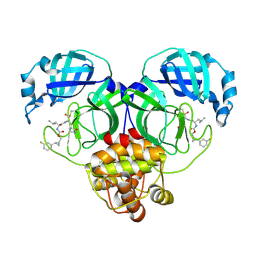 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 5c | | Descriptor: | 3C-like proteinase, N~2~-({[(1R,2R)-2-(3-fluorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ4
 
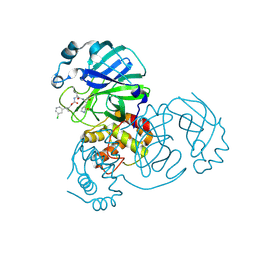 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 6c | | Descriptor: | 3C-like proteinase, N~2~-({[(1R,2R)-2-(3-chlorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7SNW
 
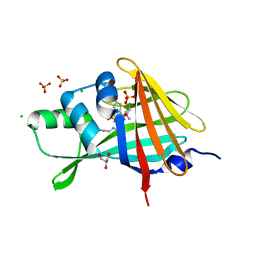 | | 1.80A Resolution Structure of NanoLuc Luciferase with Bound Inhibitor PC 16026576 | | Descriptor: | 2-(methoxycarbonyl)thiophene-3-sulfonic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.80A Resolution Structure of NanoLuc Luciferase with Bound Inhibitor PC 16026576
To be published
|
|
7SNS
 
 | | 1.55A Resolution Structure of NanoLuc Luciferase | | Descriptor: | ACETATE ION, Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55A Resolution Structure of NanoLuc Luciferase
To be published
|
|
7SNX
 
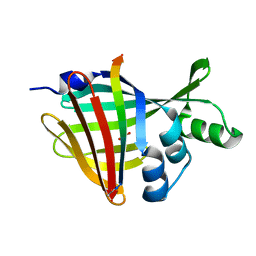 | | 1.70A Resolution Structure of NanoBiT Complementation Reporter Complex of LgBit and SmBiT Subunits | | Descriptor: | GLYCEROL, Oplophorus-luciferin 2-monooxygenase catalytic subunit, Oplophorus-luciferin 2-monooxygenase catalytic subunit: C-terminal Peptide (11-mer) | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70A Resolution Structure of NanoBiT Complementation Reporter Complex of LgBit and SmBiT Subunits
To be published
|
|
7LKT
 
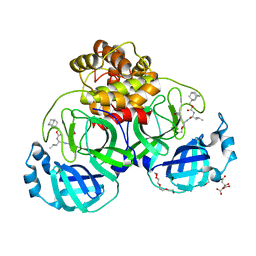 | | 1.50 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2k | | Descriptor: | (1R,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7TL7
 
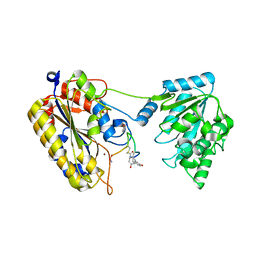 | | 1.90A resolution structure of independent phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Sa-D2) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, IMIDAZOLE, SODIUM ION, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dranchak, P, Queme, B, Aitha, M, van Neer, R.H.P, Kimura, H, Katho, T, Suga, H, Inglese, J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serum-Stable and Selective Backbone-N-Methylated Cyclic Peptides That Inhibit Prokaryotic Glycolytic Mutases.
Acs Chem.Biol., 17, 2022
|
|
7LZY
 
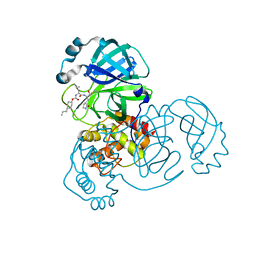 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 3c | | Descriptor: | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7M01
 
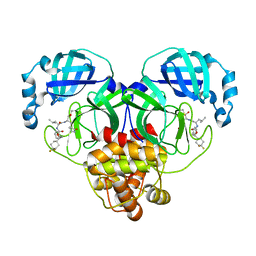 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 14c | | Descriptor: | (1R,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)-2-phenylethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)-2-phenylethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7M02
 
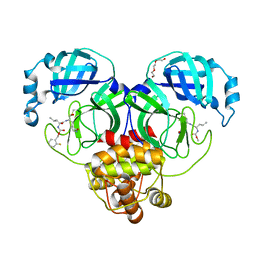 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 17c | | Descriptor: | (1R,2S)-2-((S)-2-((((2-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((2-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
