2XXA
 
 | | The Crystal Structure of the Signal Recognition Particle (SRP) in Complex with its Receptor(SR) | | Descriptor: | 4.5S RNA, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Ataide, S.F, Schmitz, N, Shen, K, Ke, A, Shan, S, Doudna, J.A, Ban, N. | | Deposit date: | 2010-11-09 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | The Crystal Structure of the Signal Recognition Particle in Complex with its Receptor.
Science, 331, 2011
|
|
6WSH
 
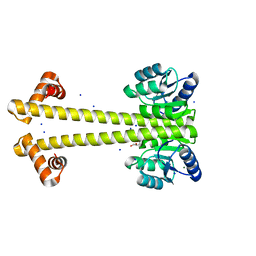 | |
6WW6
 
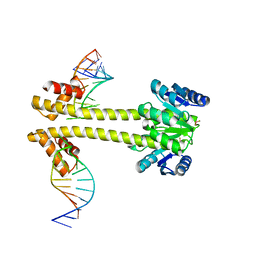 | | Crystal structure of EutV bound to RNA | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Response regulator, eutP P2 | | Authors: | Ataide, S.F, Walshe, J.L. | | Deposit date: | 2020-05-07 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural characterization of the ANTAR antiterminator domain bound to RNA.
Nucleic Acids Res., 50, 2022
|
|
6N9B
 
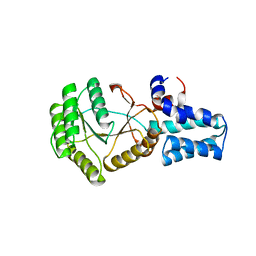 | | FtsY-NG ultra high-resolution | | Descriptor: | ACETATE ION, Signal recognition particle receptor FtsY | | Authors: | Ataide, S.F, Faoro, C. | | Deposit date: | 2018-12-02 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | Structural insights into the G-loop dynamics of E. coli FtsY NG domain.
J.Struct.Biol., 208, 2019
|
|
6N5I
 
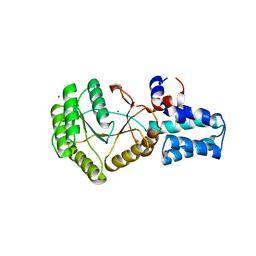 | | FtsY-NG high-resolution | | Descriptor: | GLYCEROL, POTASSIUM ION, Signal recognition particle receptor FtsY | | Authors: | Ataide, S.F, Faoro, C. | | Deposit date: | 2018-11-21 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structural insights into the G-loop dynamics of E. coli FtsY NG domain.
J.Struct.Biol., 208, 2019
|
|
6N5J
 
 | | FtsY-NG high-resolution | | Descriptor: | ACETATE ION, AMMONIUM ION, Signal recognition particle receptor FtsY | | Authors: | Ataide, S.F, Faoro, C. | | Deposit date: | 2018-11-22 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural insights into the G-loop dynamics of E. coli FtsY NG domain.
J.Struct.Biol., 208, 2019
|
|
6NC1
 
 | | FtsY-NG high-resolution | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Ataide, S.F, Faoro, C. | | Deposit date: | 2018-12-10 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Structural insights into the G-loop dynamics of E. coli FtsY NG domain.
J.Struct.Biol., 208, 2019
|
|
6NC4
 
 | | FtsY-NG high-resolution | | Descriptor: | ACETATE ION, AMMONIUM ION, GLYCEROL, ... | | Authors: | Ataide, S.F, Faoro, C. | | Deposit date: | 2018-12-10 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the G-loop dynamics of E. coli FtsY NG domain.
J.Struct.Biol., 208, 2019
|
|
6N6N
 
 | | FtsY-NG high-resolution | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ataide, S.F, Faoro, C. | | Deposit date: | 2018-11-26 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Structural insights into the G-loop dynamics of E. coli FtsY NG domain.
J.Struct.Biol., 208, 2019
|
|
4V5O
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC 40S RIBOSOMAL SUBUNIT IN COMPLEX WITH INITIATION FACTOR 1. | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S12, 40S RIBOSOMAL PROTEIN S3A, ... | | Authors: | Rabl, J, Leibundgut, M, Ataide, S.F, Haag, A, Ban, N. | | Deposit date: | 2010-11-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1.
Science, 331, 2011
|
|
8QVW
 
 | | Cryo-EM structure of the peptide binding domain of human SRP68/72 | | Descriptor: | Signal recognition particle subunit SRP68, Signal recognition particle subunit SRP72 | | Authors: | Zhong, Y, Feng, J, Koh, A.F, Kotecha, A, Greber, B.J, Ataide, S.F. | | Deposit date: | 2023-10-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of SRP68/72 reveals an extended dimerization domain with RNA-binding activity.
Nucleic Acids Res., 52, 2024
|
|
8QVX
 
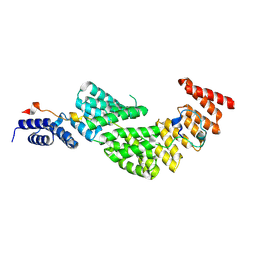 | | Structure of the PBD of human SRP68/72 (cryoSPARC 3DFlex) | | Descriptor: | Signal recognition particle subunit SRP68, Signal recognition particle subunit SRP72 | | Authors: | Zhong, Y, Feng, J, Koh, A.F, Kotecha, A, Greber, B.J, Ataide, S.F. | | Deposit date: | 2023-10-18 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | To be published
To Be Published
|
|
4C7O
 
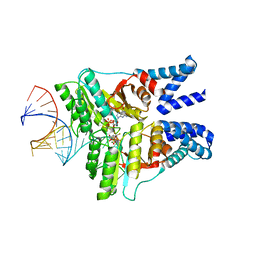 | | The structural basis of FtsY recruitment and GTPase activation by SRP RNA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SIGNAL RECOGNITION PARTICLE PROTEIN, ... | | Authors: | Voigts-Hoffmann, F, Schmitz, N, Shen, K, Shan, S.O, Ataide, S.F, Ban, N. | | Deposit date: | 2013-09-23 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis of Ftsy Recruitment and Gtpase Activation by Srp RNA
Mol.Cell, 52, 2013
|
|
2MNA
 
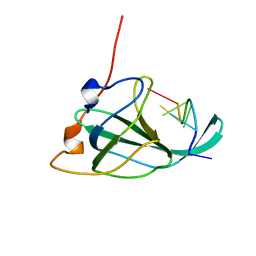 | | The structural basis of DNA binding by the single-stranded DNA-binding protein from Sulfolobus solfataricus | | Descriptor: | Single-stranded DNA binding protein (SSB), ssDNA | | Authors: | Gamsjaeger, R, Kariawasam, R, Gimenez, A.X, Touma, C.F, McIlwain, E, Bernardo, R.E, Shepherd, N.E, Ataide, S.F, Dong, A.Q, Richard, D.J, White, M.F, Cubeddu, L. | | Deposit date: | 2014-04-02 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structural basis of DNA binding by the single-stranded DNA-binding protein from Sulfolobus solfataricus
Biochem.J., 465, 2015
|
|
6CQP
 
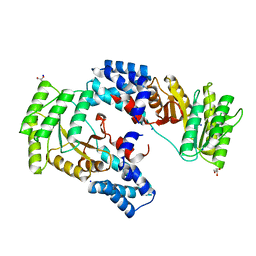 | | High resolution crystal structure of FtsY-NG domain of E. coli | | Descriptor: | GLYCEROL, SODIUM ION, Signal recognition particle receptor FtsY | | Authors: | Faoro, C, Ataide, S.F. | | Deposit date: | 2018-03-15 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | Discovery of fragments that target key interactions in the signal recognition particle (SRP) as potential leads for a new class of antibiotics.
PLoS ONE, 13, 2018
|
|
6CVD
 
 | | High resolution crystal structure of FtsY-NG domain of E. coli bound to fragment 1 | | Descriptor: | AMMONIUM ION, SODIUM ION, Signal recognition particle receptor FtsY, ... | | Authors: | Faoro, C, Ataide, S.F, Kwan, A, Wilkinson-White, L. | | Deposit date: | 2018-03-27 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of fragments that target key interactions in the signal recognition particle (SRP) as potential leads for a new class of antibiotics.
PLoS ONE, 13, 2018
|
|
6CS8
 
 | | High resolution crystal structure of FtsY-NG domain of E. coli | | Descriptor: | 1H-indole-6-carbonitrile, SODIUM ION, Signal recognition particle receptor FtsY | | Authors: | Faoro, C, Ataide, S.F. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Discovery of fragments that target key interactions in the signal recognition particle (SRP) as potential leads for a new class of antibiotics.
PLoS ONE, 13, 2018
|
|
