6NK5
 
 | |
6NK7
 
 | | Electron Cryo-Microscopy of Chikungunya in Complex with Mouse Mxra8 Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein, E1 glycoprotein, ... | | Authors: | Basore, K, Kim, A.S, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.99 Å) | | Cite: | Cryo-EM Structure of Chikungunya Virus in Complex with the Mxra8 Receptor.
Cell, 177, 2019
|
|
6NK6
 
 | | Electron Cryo-Microscopy Of Chikungunya VLP in complex with mouse Mxra8 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein, E1 glycoprotein, ... | | Authors: | Basore, K, Kim, A.S, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-22 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Cryo-EM Structure of Chikungunya Virus in Complex with the Mxra8 Receptor.
Cell, 177, 2019
|
|
2MPU
 
 | | Structural and Functional analysis of the Hordeum vulgare L. HvGR-RBP1 protein, a glycine-rich RNA binding protein implicated in the regulation of barley leaf senescence and environmental adaptation | | Descriptor: | RBP1 | | Authors: | Mason, K.E, Tripet, B.P, Eilers, B.J, Powell, P, Fischer, A.M, Copie, V. | | Deposit date: | 2014-06-02 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Biochemical Analysis of the Hordeum vulgare L. HvGR-RBP1 Protein, a Glycine-Rich RNA-Binding Protein Involved in the Regulation of Barley Plant Development and Stress Response.
Biochemistry, 53, 2014
|
|
8U65
 
 | | Cryo-EM structure of PsBphP in Pfr state, splayed PSM only | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U63
 
 | | Cryo-EM structure of PsBphP in Pfr state, Dimer of Dimers PSM only | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U64
 
 | | Cryo-EM structure of PsBphP in Pfr state, medial PSM only | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U8Z
 
 | | Cryo-EM structure of PsBphP in Pr state, extended DHp | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U4X
 
 | | Cryo-EM structure of PsBphP in Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U62
 
 | | Cryo-EM structure of PsBphP in Pfr state, Dimer of Dimers FL | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
7N1H
 
 | | CryoEM structure of Venezuelan equine encephalitis virus VLP in complex with the LDLRAD3 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Capsid, ... | | Authors: | Basore, K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-27 | | Release date: | 2021-10-13 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus in complex with the LDLRAD3 receptor.
Nature, 598, 2021
|
|
7N1I
 
 | | CryoEM structure of Venezuelan equine encephalitis virus VLP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid, E1 envelope glycoprotein, ... | | Authors: | Basore, K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-27 | | Release date: | 2021-10-13 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus in complex with the LDLRAD3 receptor.
Nature, 598, 2021
|
|
5XZE
 
 | | Mouse cGAS bound to the inhibitor RU332 | | Descriptor: | (3R)-3-[1-(3H-1lambda~4~,3-benzothiazol-2-yl)-5-hydroxy-3-methyl-1H-pyrazol-4-yl]-2-benzofuran-1(3H)-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | Authors: | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
5XZG
 
 | | Mouse cGAS bound to the inhibitor RU521 | | Descriptor: | 2-(4,5-dichloro-1H-benzimidazol-2-yl)-5-methyl-4-[(1R)-3-oxo-1,3-dihydro-2-benzofuran-1-yl]-1,2-dihydro-3H-pyrazol-3-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | Authors: | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
5XZB
 
 | | Mouse cGAS bound to the inhibitor RU365 | | Descriptor: | (3R)-3-[1-(1H-benzimidazol-2-yl)-5-hydroxy-3-methyl-1H-pyrazol-4-yl]-2-benzofuran-1(3H)-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | Authors: | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | Deposit date: | 2017-07-12 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
6QLA
 
 | | CRYSTAL STRUCTURE OF THE PMGL2 ESTERASE (point mutant 1) FROM PERMAFROST METAGENOMIC LIBRARY | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Boyko, K.M, Garsia, D, Nikolaeva, A.Y, Korzhenevskiy, D.A, Kryukova, M.V, Petrovskaya, L.E, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2019-01-31 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of PMGL2 esterase from the hormone-sensitive lipase family with GCSAG motif around the catalytic serine.
Plos One, 15, 2020
|
|
6QIN
 
 | | CRYSTAL STRUCTURE OF THE PMGL2 ESTERASE FROM PERMAFROST METAGENOMIC LIBRARY | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PMGL2 | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Kryukova, M.V, Petrovskaya, L.E, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of PMGL2 esterase from the hormone-sensitive lipase family with GCSAG motif around the catalytic serine.
Plos One, 15, 2020
|
|
6ZL7
 
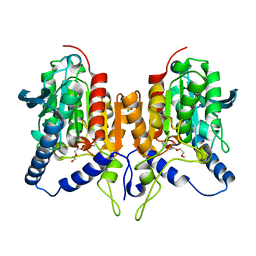 | | CRYSTAL STRUCTURE OF C173S MUTATION IN THE PMGL2 ESTERASE FROM PERMAFROST METAGENOMIC LIBRARY | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, MAGNESIUM ION, PMGL2 | | Authors: | Goryaynova, D.A, Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Kryukova, M.V, Petrovskaya, L.E, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF C173S MUTATION IN THE PMGL2 ESTERASE FROM PERMAFROST METAGENOMIC LIBRARY
To Be Published
|
|
2OYA
 
 | | Crystal structure analysis of the dimeric form of the SRCR domain of mouse MARCO | | Descriptor: | Macrophage receptor MARCO, SULFATE ION | | Authors: | Ojala, J.R.M, Pikkarainen, T, Tuuttila, A, Sandalova, T, Tryggvason, K. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition.
J.Biol.Chem., 282, 2007
|
|
2OY3
 
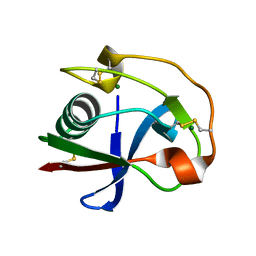 | | Crystal structure analysis of the monomeric SRCR domain of mouse MARCO | | Descriptor: | MAGNESIUM ION, Macrophage receptor MARCO | | Authors: | Ojala, J.R.M, Pikkarainen, T, Tuuttila, A, Sandalova, T, Tryggvason, K. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition.
J.Biol.Chem., 282, 2007
|
|
7B1X
 
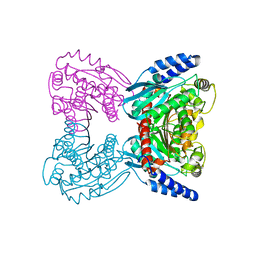 | | Crystal structure of cold-active esterase PMGL3 from permafrost metagenomic library | | Descriptor: | esterase PMGL3 | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Petrovskaya, L.E, Kryukova, M.V, Kryukova, E.A, Korzhenevsky, D.A, Lomakina, G.Y, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2020-11-25 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of a Cold-Active PMGL3 Esterase with Unusual Oligomeric Structure.
Biomolecules, 11, 2021
|
|
1EZY
 
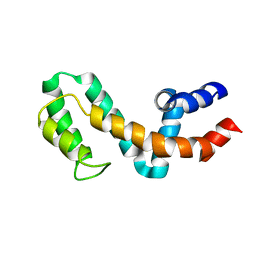 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | Descriptor: | REGULATOR OF G-PROTEIN SIGNALING 4 | | Authors: | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | Deposit date: | 2000-05-12 | | Release date: | 2001-01-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
1EZT
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | Descriptor: | REGULATOR OF G-PROTEIN SIGNALING 4 | | Authors: | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | Deposit date: | 2000-05-11 | | Release date: | 2001-01-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
1EAK
 
 | | Catalytic domain of proMMP-2 E404Q mutant | | Descriptor: | 72 KDA TYPE IV COLLAGENASE, CALCIUM ION, INHIBITOR PEPTIDE, ... | | Authors: | Bergmann, U, Tuuttila, A, Tryggvason, K, Morgunova, E. | | Deposit date: | 2001-07-12 | | Release date: | 2002-08-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal Structure of Human Mmp-2 Reveals a New P
To be Published
|
|
4NS4
 
 | | Crystal structure of cold-active estarase from Psychrobacter cryohalolentis K5T | | Descriptor: | Alpha/beta hydrolase fold protein | | Authors: | Boyko, K.M, Petrovskaya, L.E, Gorbacheva, M.A, Korgenevsky, D.A, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2013-11-28 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-dimentional structure of an esterse from Psychrobacter cryohalolentis K5T provides clues to unusual thermostability of a cold-active enzyme
To be Published
|
|
