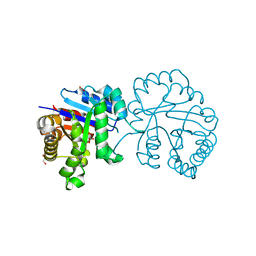
5GV4
 
 | |
5GZP
 
 | |
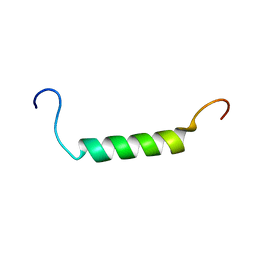
2K58
 
 | |
2K78
 
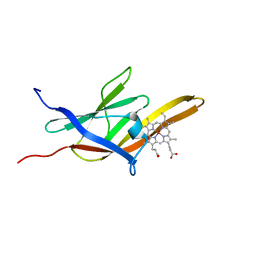 | | Solution Structure of the IsdC NEAT domain bound to Zinc Protoporphyrin | | Descriptor: | Iron-regulated surface determinant protein C, PROTOPORPHYRIN IX CONTAINING ZN | | Authors: | Villareal, V.A, Pilpa, R.M, Robson, S.A, Fadeev, E.A, Clubb, R.T. | | Deposit date: | 2008-08-06 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The IsdC Protein from Staphylococcus aureus Uses a Flexible Binding Pocket to Capture Heme.
J.Biol.Chem., 283, 2008
|
|
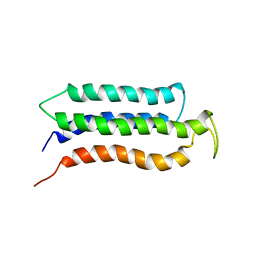
2KSR
 
 | |
2LLY
 
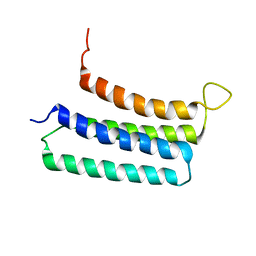 | | NMR structures of the transmembrane domains of the nAChR a4 subunit | | Descriptor: | Neuronal acetylcholine receptor subunit alpha-4 | | Authors: | Bondarenko, V, Mowrey, D, Tillman, T, Cui, T, Liu, L.T, Xu, Y, Tang, P. | | Deposit date: | 2011-11-18 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structures of the transmembrane domains of the a4b2 nAChR.
Biochim.Biophys.Acta, 1818, 2012
|
|
2LM2
 
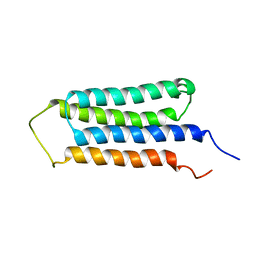 | | NMR structures of the transmembrane domains of the AChR b2 subunit | | Descriptor: | Neuronal acetylcholine receptor subunit beta-2 | | Authors: | Bondarenko, V, Mowrey, D, Tillman, T, Cui, T, Liu, L.T, Xu, Y, Tang, P. | | Deposit date: | 2011-11-18 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structures of the transmembrane domains of the a4b2 nAChR.
Biochim.Biophys.Acta, 1818, 2012
|
|
2MAW
 
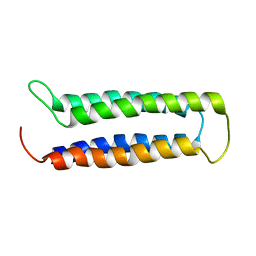 | |
1PNQ
 
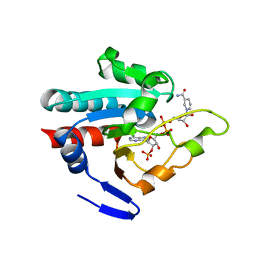 | | Crystal structure of R. rubrum transhydrogenase domain III bound to NADPH | | Descriptor: | NAD(P) transhydrogenase subunit beta, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaresan, V, Yamaguchi, M, Chartron, J, Stout, C.D. | | Deposit date: | 2003-06-12 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Change in the NADP(H) Binding Domain of Transhydrogenase Defines Four States
Biochemistry, 42, 2003
|
|
1PNO
 
 | | Crystal structure of R. rubrum transhydrogenase domain III bound to NADP | | Descriptor: | NAD(P) transhydrogenase subunit beta, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaresan, V, Yamaguchi, M, Chartron, J, Stout, C.D. | | Deposit date: | 2003-06-12 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Change in the NADP(H) Binding Domain of Transhydrogenase Defines Four States
Biochemistry, 42, 2003
|
|
6YXB
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant (space group P21) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SULFATE ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
6YX6
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant (no morpholine) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SULFATE ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
6YX4
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant with morpholine | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
6YWI
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148E variant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YWH
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148D variant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YXC
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148R variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SODIUM ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YWQ
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148H variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YWR
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148H variant (space group C2) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SULFATE ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YWG
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148N variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
5FCH
 
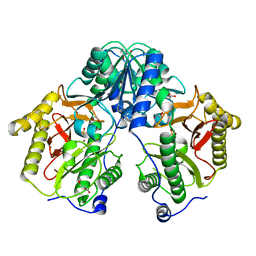 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris, phosphate and Zn bound | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLY-GLY-GLY, GLYCEROL, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and biochemical investigations reveal novel mode of substrate selectivity and illuminate substrate inhibition and allostericity in a subfamily of Xaa-Pro dipeptidases
Biochim. Biophys. Acta, 1865, 2017
|
|
5FCF
 
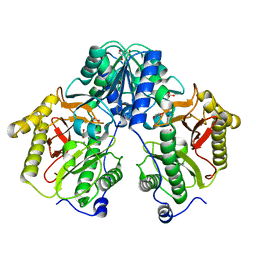 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris, phosphate and Mn bound | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLY-GLY-GLY, GLYCEROL, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and biochemical investigations reveal novel mode of substrate selectivity and illuminate substrate inhibition and allostericity in a subfamily of Xaa-Pro dipeptidases.
Biochim. Biophys. Acta, 1865, 2017
|
|
6A8M
 
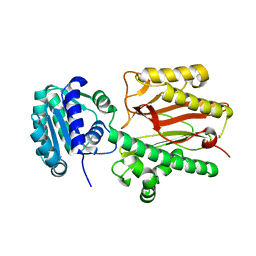 | | N-terminal domain of FACT complex subunit SPT16 from Eremothecium gossypii (Ashbya gossypii) | | Descriptor: | FACT complex subunit SPT16 | | Authors: | Gaur, N.K, Are, V.N, Durani, V, Ghosh, B, Kumar, A, Kulkarni, K, Makde, R.D. | | Deposit date: | 2018-07-09 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolutionary conservation of protein dynamics: insights from all-atom molecular dynamics simulations of 'peptidase' domain of Spt16.
J.Biomol.Struct.Dyn., 2021
|
|
5CNX
 
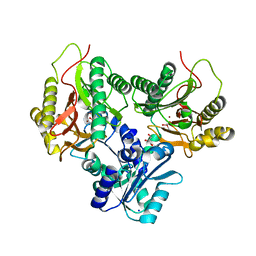 | | Crystal structure of Xaa-Pro aminopeptidase from Escherichia coli K12 | | Descriptor: | Aminopeptidase YpdF, CACODYLATE ION, GLYCEROL, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-07-18 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures and activities of widely conserved small prokaryotic aminopeptidases-P clarify classification of M24B peptidases
Proteins, 2018
|
|
4R60
 
 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Proline dipeptidase, ... | | Authors: | Kumar, A, Ghosh, B, Are, V.N, Jamdar, S.N, Makde, R.D, Sharma, S.M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris
to be published
|
|
5CDE
 
 | | R372A mutant of Xaa-Pro dipeptidase from Xanthomonas campestris | | Descriptor: | Proline dipeptidase, SULFATE ION, ZINC ION | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R. | | Deposit date: | 2015-07-03 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | R372A mutant of Xaa-Pro dipeptidase from Xanthomonas campestris at 1.85 Angstrom resolution
To Be Published
|
|
