6Y7U
 
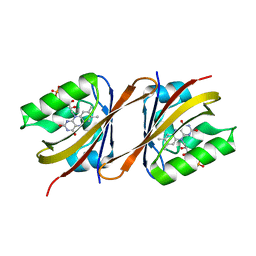 | | Structure of Chloroflexus aggregans Cagg_3753 LOV domain C85A A95P variant (CagFbFP) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gordeliy, V, Gushchin, I. | | Deposit date: | 2020-03-02 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Effects of Proline Substitutions on the Thermostable LOV Domain from Chloroflexus aggregans
Crystals, 2020
|
|
6YXB
 
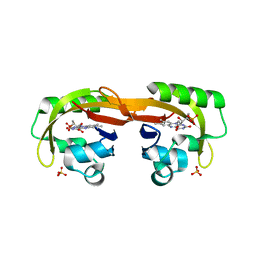 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant (space group P21) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SULFATE ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
6YX6
 
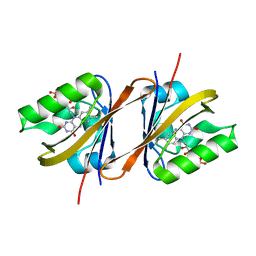 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant (no morpholine) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SULFATE ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
6YX4
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant with morpholine | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
7NE4
 
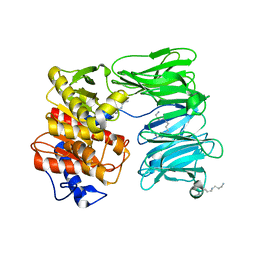 | | E125A mutant of oligopeptidase B from S. proteomaculans with modified hinge region | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | First Crystal Structure of Bacterial Oligopeptidase B in an Intermediate State: The Roles of the Hinge Region Modification and Spermine.
Biology (Basel), 10, 2021
|
|
7QZV
 
 | | Hm-AMP2 | | Descriptor: | Hm-AMP2 | | Authors: | Grafskaia, E.N, Pavlova, E.R, Latsis, I.A, Malakhova, M.V, Lavrenova, V.N, Ivchenkov, D.V, Bashkirov, P.V, Kot, E.F, Mineev, K.S, Arseniev, A.S, Klinov, D.V, Lazarev, V.N. | | Deposit date: | 2022-02-01 | | Release date: | 2022-11-23 | | Method: | SOLUTION NMR | | Cite: | Non-toxic antimicrobial peptide Hm-AMP2 from leech metagenome proteins identified by the gradient-boosting approach
Materials, 224, 2022
|
|
7QZW
 
 | | Hm-AMP8 | | Descriptor: | Hm-AMP8 | | Authors: | Grafskaia, E.N, Pavlova, E.R, Latsis, I.A, Malakhova, M.V, Lavrenova, V.N, Ivchenkov, D.V, Bashkirov, P.V, Kot, E.F, Mineev, K.S, Arseniev, A.S, Klinov, D.V, Lazarev, V.N. | | Deposit date: | 2022-02-01 | | Release date: | 2023-02-15 | | Method: | SOLUTION NMR | | Cite: | The study of the interaction of cationic peptides from a medical leech with blood components as a first step to the systemic use of antimicrobial peptides for therapy of infectious diseases
To Be Published
|
|
7NE5
 
 | | catalytically non active S532A mutant of oligopeptidase B from S. proteomaculans with modified hinge region | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | First Crystal Structure of Bacterial Oligopeptidase B in an Intermediate State: The Roles of the Hinge Region Modification and Spermine.
Biology (Basel), 10, 2021
|
|
7OB1
 
 | | OLIGOPEPTIDASE B FROM S. PROTEOMACULANS WITH MODIFIED HINGE | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Korzhenevskiy, D.A, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | First Crystal Structure of Bacterial Oligopeptidase B in an Intermediate State: The Roles of the Hinge Region Modification and Spermine.
Biology (Basel), 10, 2021
|
|
6YWI
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148E variant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YWH
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148D variant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YXC
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148R variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SODIUM ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YWQ
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148H variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YWR
 
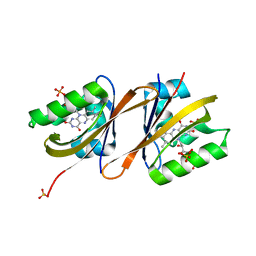 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148H variant (space group C2) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SULFATE ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YWG
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148N variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
7AB7
 
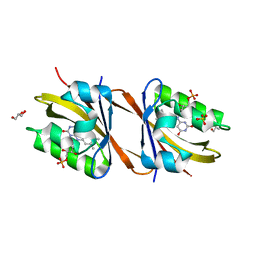 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) I52T Q148K variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-09-06 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
7AB6
 
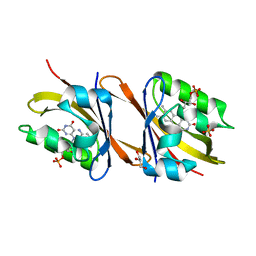 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) I52T variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-09-06 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
7NE7
 
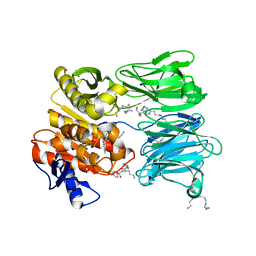 | | oligopeptidase B from S. proteomaculans with modified hinge region in complex with N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide | | Descriptor: | N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2021-02-03 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of N alpha-p-tosyl-lysyl Chloromethylketone-Bound Oligopeptidase B from Serratia Proteamaculans Revealed a New Type of Inhibitor Binding
Crystals, 11, 2021
|
|
6W2S
 
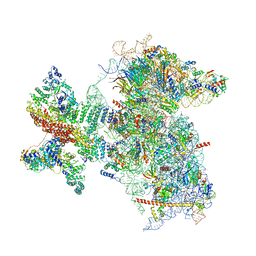 | | Structure of the Cricket Paralysis Virus 5-UTR IRES (CrPV 5-UTR-IRES) bound to the small ribosomal subunit in the open state (Class 1) | | Descriptor: | 18S rRNA, CrPV 5'-UTR IRES, Eukaryotic translation initiation factor 3 subunit A, ... | | Authors: | Neupane, R, Pisareva, V, Rodriguez, C.F, Pisarev, A, Fernandez, I.S. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | A complex IRES at the 5'-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate.
Elife, 9, 2020
|
|
6QWX
 
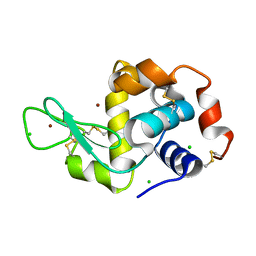 | | HEWL lysozyme, crystallized from NiCl2 solution | | Descriptor: | CHLORIDE ION, Lysozyme C, NICKEL (II) ION | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
2M61
 
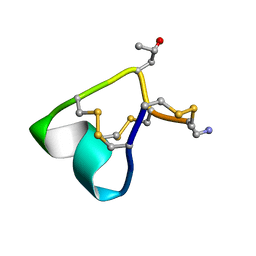 | | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails | | Descriptor: | Conotoxin Ar1446 | | Authors: | Sarma, S.P, Rajesh, R.P, Kumar, G.S, Sudarslal, S, Sabareesh, V, Gowd, K.H, Gupta, K, Krishnan, K.S, Balaram, P. | | Deposit date: | 2013-03-18 | | Release date: | 2014-04-16 | | Method: | SOLUTION NMR | | Cite: | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails
To be Published
|
|
2JST
 
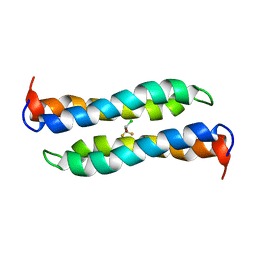 | | Four-Alpha-Helix Bundle with Designed Anesthetic Binding Pockets II: Halothane Effects on Structure and Dynamics | | Descriptor: | 2-BROMO-2-CHLORO-1,1,1-TRIFLUOROETHANE, Four-Alpha-Helix Bundle | | Authors: | Cui, T, Bondarenko, V, Ma, D, Canlas, C, Brandon, N.R, Johansson, J.S, Tang, P, Xu, Y. | | Deposit date: | 2007-07-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Four-alpha-helix bundle with designed anesthetic binding pockets. Part II: halothane effects on structure and dynamics
Biophys.J., 94, 2008
|
|
6QWW
 
 | | HEWL lysozyme, crystallized from CuCl2 solution | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Lysozyme C | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
6QWY
 
 | | HEWL lysozyme, crystallized from NaCl solution | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
6QWZ
 
 | | HEWL lysozyme, crystallized from KCl solution | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
