3ACZ
 
 | | Crystal structure of Entamoeba histolytica methionine gamma-lyase 1 | | Descriptor: | GLYCEROL, Methionine gamma-lyase, SULFATE ION | | Authors: | Karaki, T, Sato, D, Shimizu, A, Nozaki, T, Harada, S. | | Deposit date: | 2010-01-13 | | Release date: | 2011-02-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of Entamoeba histolytica methionine gamma-lyase 1
To be Published
|
|
3AEN
 
 | | Reaction intermediate structure of Entamoeba histolytica methionine gamma-lyase 1 containing Michaelis complex and alpha-amino-alpha, beta-butenoic acid-pyridoxal-5'-phosphate | | Descriptor: | (2E)-2-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}but-2-enoic acid, GLYCEROL, METHIONINE, ... | | Authors: | Karaki, T, Sato, D, Shimizu, A, Nozaki, T, Harada, S. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Entamoeba histolytica methionine gamma-lyase 1
To be Published
|
|
3AEM
 
 | | Reaction intermediate structure of Entamoeba histolytica methionine gamma-lyase 1 containing Michaelis complex and methionine imine-pyridoxamine-5'-phosphate | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-4-(methylsulfanyl)butanoic acid, GLYCEROL, METHIONINE, ... | | Authors: | Karaki, T, Sato, D, Shimizu, A, Nozaki, T, Harada, S. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Entamoeba histolytica methionine gamma-lyase 1
To be published
|
|
3AEL
 
 | | Reaction intermediate structure of Entamoeba histolytica methionine gamma-lyase 1 containing methionine imine-pyridoxamine-5'-phosphate and alpha-amino-alpha, beta-butenoic acid-pyridoxal-5'-phosphate | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-4-(methylsulfanyl)butanoic acid, (2E)-2-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}but-2-enoic acid, GLYCEROL, ... | | Authors: | Karaki, T, Sato, D, Shimizu, A, Nozaki, T, Harada, S. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Entamoeba histolytica methionine gamma-lyase 1
To be Published
|
|
3AEJ
 
 | | Reaction intermediate structure of Entamoeba histolytica methionine gamma-lyase 1 tetramer containing Michaelis complex and methionine-pyridoxal-5'-phosphate | | Descriptor: | GLYCEROL, METHIONINE, Methionine gamma-lyase, ... | | Authors: | Karaki, T, Sato, D, Shimizu, A, Nozaki, T, Harada, S. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of Entamoeba histolytica methionine gamma-lyase 1
To be Published
|
|
3AEP
 
 | | Reaction intermediate structure of Entamoeba histolytica methionine gamma-lyase 1 containing alpha-amino-alpha, beta-butenoic acid-pyridoxal-5'-phosphate | | Descriptor: | (2E)-2-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}but-2-enoic acid, GLYCEROL, METHANETHIOL, ... | | Authors: | Karaki, T, Sato, D, Shimizu, A, Nozaki, T, Harada, S. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of Entamoeba histolytica methionine gamma-lyase 1
To be Published
|
|
3AEO
 
 | | Reaction intermediate structure of Entamoeba histolytica methionine gamma-lyase 1 containing methionine alpha, beta-enamine-pyridoxamine-5'-phosphate | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-4-(methylsulfanyl)but-2-enoic acid, GLYCEROL, Methionine gamma-lyase, ... | | Authors: | Karaki, T, Sato, D, Shimizu, A, Nozaki, T, Harada, S. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of Entamoeba histolytica methionine gamma-lyase 1
To be Published
|
|
4PJ2
 
 | | Crystal structure of Aeromonas hydrophila PliI in complex with Meretrix lusoria lysozyme | | Descriptor: | GLYCEROL, Lysozyme, MAGNESIUM ION, ... | | Authors: | Leysen, S, Van Herreweghe, J.M, Yoneda, K, Ogata, M, Usui, T, Michiels, C.W, Araki, T, Strelkov, S.V. | | Deposit date: | 2014-05-10 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The structure of the proteinaceous inhibitor PliI from Aeromonas hydrophila in complex with its target lysozyme.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5B1Y
 
 | | Crystal structure of NADPH bound carbonyl reductase from Aeropyrum pernix | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yoneda, K, Sakuraba, H, Fukuda, Y, Araki, T, Ohshima, T. | | Deposit date: | 2015-12-22 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Catalytic properties and crystal structure of thermostable NAD(P)H-dependent carbonyl reductase from the hyperthermophilic archaeon Aeropyrum pernix K1.
Enzyme.Microb.Technol., 91, 2016
|
|
6K5J
 
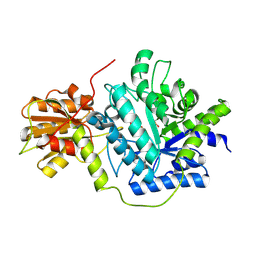 | | Structure of a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GH3 beta-N-acetylglucosaminidase, GLYCEROL | | Authors: | Itoh, T, Araki, T, Nishiyama, T, Hibi, T, Kimoto, H. | | Deposit date: | 2019-05-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structural and functional characterization of a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7.
J.Biochem., 166, 2019
|
|
2COV
 
 | | Crystal structure of CBM31 from beta-1,3-xylanase | | Descriptor: | beta-1,3-xylanase | | Authors: | Hashimoto, H, Tamai, Y, Okazaki, F, Tamaru, Y, Shimizu, T, Araki, T, Sato, M. | | Deposit date: | 2005-05-18 | | Release date: | 2005-09-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The first crystal structure of a family 31 carbohydrate-binding module with affinity to beta-1,3-xylan
Febs Lett., 579, 2005
|
|
3AYQ
 
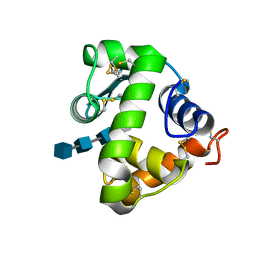 | | Crystal structure of inhibitor bound lysozyme from Meretrix lusoria | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-[(5S,6R)-5-hydroxy-6-(hydroxymethyl)-2-oxo-5,6-dihydro-2H-pyran-3-yl]acetamide, Lysozyme | | Authors: | Yoneda, K, Kuwano, Y, Usui, T, Ogata, M, Suzuki, A, Araki, T. | | Deposit date: | 2011-05-13 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of inhibitor bound lysozyme from Meretrix lusoria
To be Published
|
|
2ZPO
 
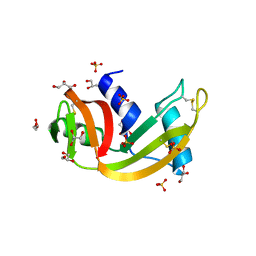 | | Crystal Structure of Green Turtle Egg White Ribonuclease | | Descriptor: | GLYCEROL, Ribonuclease, SULFATE ION | | Authors: | Katekaew, S, Kakuta, Y, Torikata, T, Kimura, M, Yoneda, K, Araki, T. | | Deposit date: | 2008-07-25 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Newly Found Green Turtle Egg White Ribonuclease
To be Published
|
|
3WXB
 
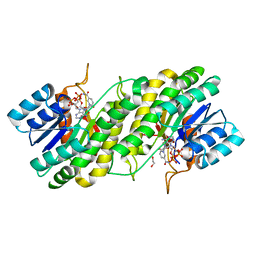 | | Crystal structure of NADPH bound carbonyl reductase from chicken fatty liver | | Descriptor: | 1,2-ETHANEDIOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized protein | | Authors: | Yoneda, K, Sakuraba, H, Fukuda, Y, Sone, T, Araki, T, Ohshima, T. | | Deposit date: | 2014-07-29 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A novel NAD(P)H-dependent carbonyl reductase specifically expressed in the thyroidectomized chicken fatty liver: catalytic properties and crystal structure.
Febs J., 282, 2015
|
|
3AB6
 
 | | Crystal structure of NAG3 bound lysozyme from Meretrix lusoria | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme | | Authors: | Yoneda, K, Kuwano, Y, Araki, T. | | Deposit date: | 2009-12-01 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The tertiary structure of an i-type lysozyme isolated from the common orient clam (Meretrix lusoria)
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3WYH
 
 | |
3WVX
 
 | |
3WVY
 
 | | Structure of D48A hen egg white lysozyme in complex with (GlcNAc)4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme C | | Authors: | Kawaguchi, Y, Yoneda, K, Araki, T. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The role of Asp48 in the hydrogen bonding network involving Asp52 of hen egg white lysozyme
TO BE PUBLISHED
|
|
