7P43
 
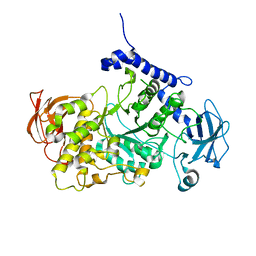 | | Structure of CgGBE in complex with maltotriose | | Descriptor: | 1,4-alpha-glucan-branching enzyme, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ballut, L, Conchou, L, Violot, S, Galisson, F, Aghajari, N. | | Deposit date: | 2021-07-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Candida glabrata glycogen branching enzyme structure reveals unique features of branching enzymes of the Saccharomycetaceae phylum.
Glycobiology, 32, 2022
|
|
7P44
 
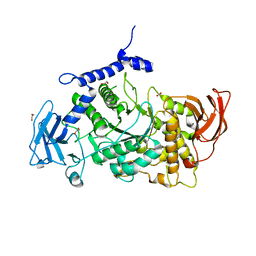 | | Structure of CgGBE in P21212 space group | | Descriptor: | 1,2-ETHANEDIOL, 1,4-alpha-glucan-branching enzyme | | Authors: | Ballut, L, Conchou, L, Violot, S, Galisson, F, Aghajari, N. | | Deposit date: | 2021-07-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Candida glabrata glycogen branching enzyme structure reveals unique features of branching enzymes of the Saccharomycetaceae phylum.
Glycobiology, 32, 2022
|
|
7P45
 
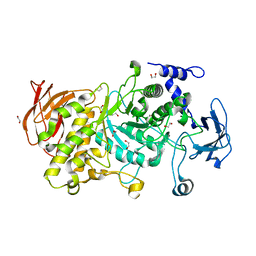 | | Structure of CgGBE in P212121 space group | | Descriptor: | 1,2-ETHANEDIOL, 1,4-alpha-glucan-branching enzyme | | Authors: | Ballut, L, Conchou, L, Violot, S, Galisson, F, Aghajari, N. | | Deposit date: | 2021-07-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Candida glabrata glycogen branching enzyme structure reveals unique features of branching enzymes of the Saccharomycetaceae phylum.
Glycobiology, 32, 2022
|
|
1K72
 
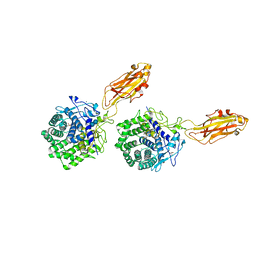 | | The X-ray Crystal Structure Of Cel9G Complexed With cellotriose | | Descriptor: | CALCIUM ION, Endoglucanase 9G, GLYCEROL, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2001-10-18 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
1KFW
 
 | |
1O0Q
 
 | | Crystal structure of a cold adapted alkaline protease from Pseudomonas TAC II 18, co-crystallized with 1 mM EDTA | | Descriptor: | CALCIUM ION, SULFATE ION, serralysin | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-24 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
1O0T
 
 | | CRYSTAL STRUCTURE OF A COLD ADAPTED ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18, CO-CRYSTALLIZED WITH 5 mM EDTA (5 DAYS) | | Descriptor: | CALCIUM ION, SULFATE ION, serralysin | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-24 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
1OM6
 
 | | CRYSTAL STRUCTURE OF A COLD ADAPTED ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18, CO-CRYSTALLIZED WITH 5mM EDTA (2 MONTHS) | | Descriptor: | CALCIUM ION, SULFATE ION, serralysin | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
1OM8
 
 | | CRYSTAL STRUCTURE OF A COLD ADAPTED ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18, CO-CRYSTALLYZED WITH 10 mM EDTA | | Descriptor: | CALCIUM ION, SERRALYSIN, SULFATE ION | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
1OM7
 
 | | CRYSTAL STRUCTURE OF A COLD ADAPTED ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18, SOAKED IN 85 mM EDTA | | Descriptor: | CALCIUM ION, SERRALYSIN, SULFATE ION | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
1OMJ
 
 | | CRYSTAL STRUCTURE OF A PSYCHROPHILIC ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18 | | Descriptor: | CALCIUM ION, SERRALYSIN, SULFATE ION, ... | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
4D5U
 
 | | Structure of OmpF in I2 | | Descriptor: | OUTER MEMBRANE PROTEIN F | | Authors: | Chaptal, V, Kilburg, A, Flot, D, Wiseman, B, Aghajari, N, Jault, J.M, Falson, P. | | Deposit date: | 2014-11-07 | | Release date: | 2015-12-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Two Different Centered Monoclinic Crystals of the E. Coli Outer-Membrane Protein Ompf Originate from the Same Building Block.
Biochim.Biophys.Acta, 1858, 2016
|
|
4GIA
 
 | | Crystal structure of the MUTB F164L mutant from crystals soaked with isomaltulose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GI9
 
 | | Crystal structure of the MUTB F164L mutant from crystals soaked with Trehalulose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GI8
 
 | | Crystal structure of the MUTB F164L mutant from crystals soaked with the substrate sucrose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GIN
 
 | | Crystal structure of the MUTB R284C mutant from crystals soaked with the inhibitor deoxynojirimycin | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase | | Authors: | Lipski, A, Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GI6
 
 | | Crystal structure of the MUTB F164L mutant in complex with glucose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GO9
 
 | | CRYSTAL STRUCTURE of the TREHALULOSE SYNTHASE MUTANT, MUTB D415N, in COMPLEX with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Sucrose isomerase | | Authors: | Lipski, A, Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE of the TREHALULOSE SYNTHASE MUTB, MUTANT D415N, in COMPLEX with TRIS
To be Published, 2013
|
|
4GO8
 
 | | Crystal Structure of the TREHALULOSE SYNTHASE MUTB, MUTANT A258V, in complex with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Sucrose isomerase | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of MUTB A258V mutant in complex with TRIS
To be Published, 2013
|
|
4H8H
 
 | | MUTB inactive double mutant E254Q-D415N | | Descriptor: | CALCIUM ION, GLYCEROL, SULFATE ION, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-09-22 | | Release date: | 2013-09-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into product binding in sucrose isomerases from crystal structures of MutB from Rhizobium sp.
To be Published
|
|
4H8V
 
 | |
4H7V
 
 | |
4H4B
 
 | | Human cytosolic 5'-nucleotidase II in complex with Anthraquinone-2,6- disulfonic acid | | Descriptor: | 9,10-dioxo-9,10-dihydroanthracene-2,6-disulfonic acid, Cytosolic purine 5'-nucleotidase, GLYCEROL, ... | | Authors: | Rhimi, M, Aghajari, N. | | Deposit date: | 2012-09-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification and characterization of inhibitors of cytoplasmic 5'-nucleotidase cN-II issued from virtual screening.
Biochem Pharmacol, 85, 2013
|
|
4H8U
 
 | |
4H2C
 
 | | Trehalulose synthase MutB R284C mutant | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase | | Authors: | Lipski, A, Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2012-09-12 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
