7P45
 
 | | Structure of CgGBE in P212121 space group | | Descriptor: | 1,2-ETHANEDIOL, 1,4-alpha-glucan-branching enzyme | | Authors: | Ballut, L, Conchou, L, Violot, S, Galisson, F, Aghajari, N. | | Deposit date: | 2021-07-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Candida glabrata glycogen branching enzyme structure reveals unique features of branching enzymes of the Saccharomycetaceae phylum.
Glycobiology, 32, 2022
|
|
7O7T
 
 | | Structure of the PL6 family alginate lyase Patl3640 from Pseudoalteromonas atlantica T6c in complex with 4-deoxy-L-erythro-5-hexoseulose uronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-L-erythro-hex-5-ulosuronic acid, GLYCEROL, ... | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O84
 
 | | Structure of the PL6 family alginate lyase Pedsa0632 from Pseudopedobacter saltans in complex with substrate | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, Alginate lyase | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O77
 
 | | Structure of the PL6 family alginate lyase Patl3640 from Pseudoalteromonas atlantica T6c | | Descriptor: | GLYCEROL, Poly(Beta-D-mannuronate) lyase | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.321 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O7A
 
 | |
7O78
 
 | | Structure of the PL6 family chondroitinase B from Pseudopedobacter saltans, Pedsa3807 | | Descriptor: | Polysaccharide lyase from Pseudopedobacter saltans, Pedsa3807 | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O79
 
 | | Structure of the PL6 family polysaccharide lyase Pedsa3628 from Pseudopedobacter saltans | | Descriptor: | PHOSPHATE ION, Poly(Beta-D-mannuronate) lyase | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
3GBE
 
 | | Crystal structure of the isomaltulose synthase SmuA from Protaminobacter rubrum in complex with the inhibitor deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, CITRATE ANION, ... | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2009-02-19 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural determinants of product specificity of sucrose isomerases
Febs Lett., 583, 2009
|
|
3GBD
 
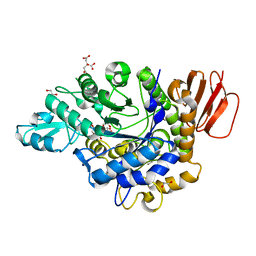 | | Crystal structure of the isomaltulose synthase SmuA from Protaminobacter rubrum | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Sucrose isomerase SmuA from Protaminobacter rubrum | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2009-02-19 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural determinants of product specificity of sucrose isomerases
Febs Lett., 583, 2009
|
|
8R3S
 
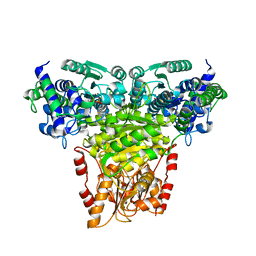 | | Transketolase from Staphylococcus aureus in complex with thiamin pyrophosphate | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, Transketolase | | Authors: | Ballut, L, Georges, N, Aghajari, N, Hecquet, L, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-11-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Are Transketolases relevant Targets fighting Human Pathogens? A comparative Biochemical, Bioinformatic and Structural Study
To Be Published
|
|
8R3O
 
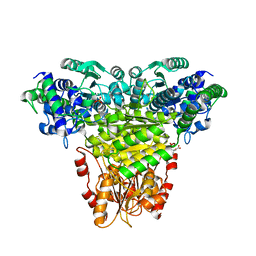 | | Transketolase from Haemophilus influenzae in complex with thiamin pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Ballut, L, Georges, R.N, Aghajari, N, Hecquet, L, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-11-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Are Transketolases relevant Targets fighting Human Pathogens? A comparative Biochemical, Bioinformatic and Structural Study
To Be Published
|
|
8R3R
 
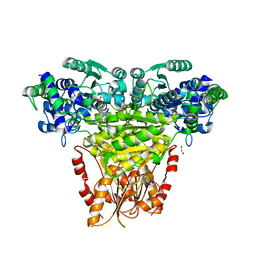 | | Transketolase from Streptococcus pneumoniae in complex with thiamin pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable transketolase, ... | | Authors: | Ballut, L, Georges, R.N, Aghajari, N, Hecquet, L, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-11-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Are Transketolases relevant Targets fighting Human Pathogens? A comparative Biochemical, Bioinformatic and Structural Study
To Be Published
|
|
8R3P
 
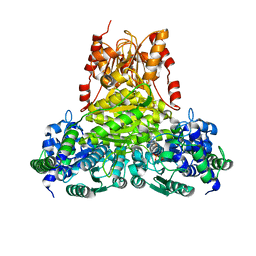 | | Transketolase from Enterococcus faecium in complex with thiamin pyrophosphate | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, Transketolase | | Authors: | Ballut, L, Georges, R.N, Aghajari, N, Hecquet, L, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-11-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Are Transketolases relevant Targets fighting Human Pathogens? A comparative Biochemical, Bioinformatic and Structural Study
To Be Published
|
|
8R3Q
 
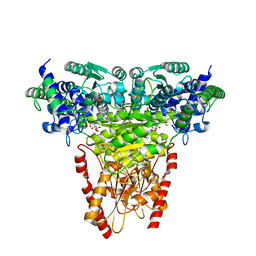 | | Transketolase from Plasmodium falciparum in complex with thiamin pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Ballut, L, Georges, R.N, Aghajari, N, Hecquet, L, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-11-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Are Transketolases relevant Targets fighting Human Pathogens? A comparative Biochemical, Bioinformatic and Structural Study
To Be Published
|
|
1G87
 
 | | THE CRYSTAL STRUCTURE OF ENDOGLUCANASE 9G FROM CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOCELLULASE 9G, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2000-11-16 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
6HXE
 
 | |
1GA2
 
 | | THE CRYSTAL STRUCTURE OF ENDOGLUCANASE 9G FROM CLOSTRIDIUM CELLULOLYTICUM COMPLEXED WITH CELLOBIOSE | | Descriptor: | ACETIC ACID, CALCIUM ION, ENDOGLUCANASE 9G, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2000-11-29 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
6I06
 
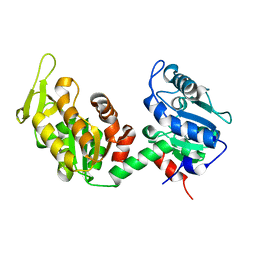 | |
1HT6
 
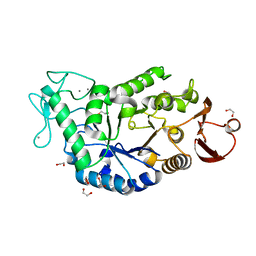 | | CRYSTAL STRUCTURE AT 1.5A RESOLUTION OF THE BARLEY ALPHA-AMYLASE ISOZYME 1 | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-AMYLASE ISOZYME 1, CALCIUM ION | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2000-12-29 | | Release date: | 2003-07-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of barley alpha-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: a pair of sugar tongs
Structure, 11, 2003
|
|
1H71
 
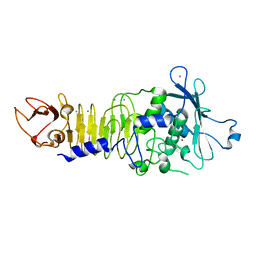 | | Psychrophilic Protease from Pseudoalteromonas 'TAC II 18' | | Descriptor: | CALCIUM ION, SERRALYSIN, ZINC ION | | Authors: | Villeret, V, Van Petegem, F, Aghajari, N, Chessa, J.-P, Gerday, C, Haser, R, Van Beeumen, J. | | Deposit date: | 2001-07-02 | | Release date: | 2003-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of a Psychrophilic Metalloprotease Reveal New Insights Into Catalysis by Cold-Adapted Proteases
Proteins: Struct.,Funct., Genet., 50, 2003
|
|
2PWD
 
 | | Crystal Structure of the Trehalulose Synthase MUTB from Pseudomonas Mesoacidophila MX-45 Complexed to the Inhibitor Deoxynojirmycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, CALCIUM ION, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
2PWF
 
 | | Crystal structure of the MutB D200A mutant in complex with glucose | | Descriptor: | CALCIUM ION, Sucrose isomerase, beta-D-glucopyranose | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
2PWG
 
 | | Crystal Structure of the Trehalulose Synthase MutB From Pseudomonas Mesoacidophila MX-45 Complexed to the Inhibitor Castanospermine | | Descriptor: | CALCIUM ION, CASTANOSPERMINE, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
2PWE
 
 | | Crystal structure of the MutB E254Q mutant in complex with the substrate sucrose | | Descriptor: | CALCIUM ION, Sucrose isomerase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 282, 2007
|
|
2PWH
 
 | | Crystal structure of the trehalulose synthase MutB from Pseudomonas mesoacidophila MX-45 | | Descriptor: | CALCIUM ION, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
