3VTJ
 
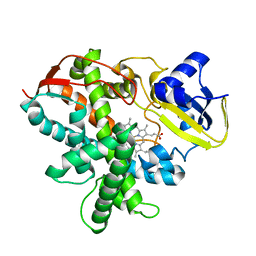 | | Cytochrome P450SP alpha (CYP152B1) mutant A245H | | Descriptor: | Fatty acid alpha-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fujishiro, T, Shoji, O, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2012-05-30 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A substrate-binding-state mimic of H2O2-dependent cytochrome P450 produced by one-point mutagenesis and peroxygenation of non-native substrates
Catalysis Science And Technology, 6, 2016
|
|
2ZQX
 
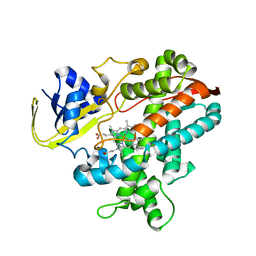 | | Cytochrome P450BSbeta cocrystallized with heptanoic acid | | Descriptor: | Cytochrome P450 152A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shoji, O, Fujishiro, T, Nagano, S, Hirose, T, Shiro, Y, Watanabe, Y. | | Deposit date: | 2008-08-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Understanding substrate misrecognition of hydrogen peroxide dependent cytochrome P450 from Bacillus subtilis.
J.Biol.Inorg.Chem., 2010
|
|
6J9X
 
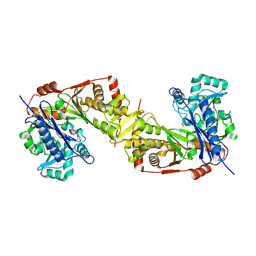 | | Crystal structure of Trypanosoma brucei gambiense glycerol kinase phosphorylated at Thr12(pyrophosphatase reaction) | | Descriptor: | GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Chishima, T, Ichinose, M, Inaoka, D.K, Kido, Y, Ibrahim, B, Bringaud, F, de Koning, H, McKerrow, J.H, Watanabe, Y, Nozaki, T, Michels, P.A.M, Harada, S, Kita, K, Shiba, T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Glycerol Kinase of African Human Trypanosomes Possesses a Pyrophosphatase Activity.
To Be Published
|
|
2ZQJ
 
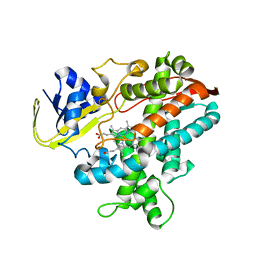 | | Substrate-Free Form of Cytochrome P450BSbeta | | Descriptor: | Cytochrome P450 152A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shoji, O, Fujishiro, T, Nagano, S, Hirose, T, Shiro, Y, Watanabe, Y. | | Deposit date: | 2008-08-11 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding substrate misrecognition of hydrogen peroxide dependent cytochrome P450 from Bacillus subtilis.
J.Biol.Inorg.Chem., 2010
|
|
3AWM
 
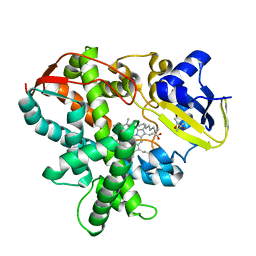 | | Cytochrome P450SP alpha (CYP152B1) wild-type with palmitic acid | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Fatty acid alpha-hydroxylase, PALMITIC ACID, ... | | Authors: | Fujishiro, T, Shoji, O, Nagano, S, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of H2O2-dependent cytochrome P450SPalpha with its bound fatty acid substrate: insight into the regioselective hydroxylation of fatty acids at the alpha position
J.Biol.Chem., 286, 2011
|
|
3AWP
 
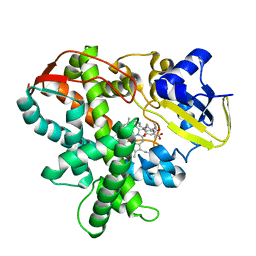 | | Cytochrome P450SP alpha (CYP152B1) mutant F288G | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Fatty acid alpha-hydroxylase, PALMITIC ACID, ... | | Authors: | Fujishiro, T, Shoji, O, Nagano, S, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of H2O2-dependent cytochrome P450SPalpha with its bound fatty acid substrate: insight into the regioselective hydroxylation of fatty acids at the alpha position.
J.Biol.Chem., 286, 2011
|
|
3AWQ
 
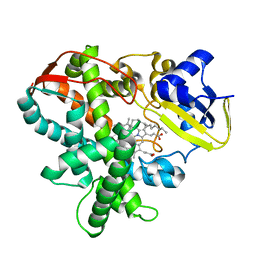 | | Cytochrome P450SP alpha (CYP152B1) mutant L78F | | Descriptor: | Fatty acid alpha-hydroxylase, PALMITIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fujishiro, T, Shoji, O, Nagano, S, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of H2O2-dependent cytochrome P450SPalpha with its bound fatty acid substrate: insight into the regioselective hydroxylation of fatty acids at the alpha position.
J.Biol.Chem., 286, 2011
|
|
7Y9P
 
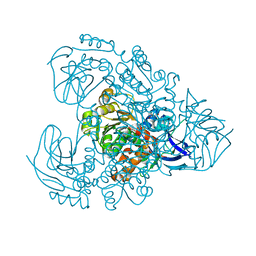 | | Xylitol dehydrogenase S96C/S99C/Y102C mutant(thermostabilized form) from Pichia stipitis | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Yoshiwara, K, Watanabe, Y, Watanabe, S. | | Deposit date: | 2022-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular evolutionary insight of structural zinc atom in yeast xylitol dehydrogenases and its application in bioethanol production by lignocellulosic biomass.
Sci Rep, 13, 2023
|
|
2Z6B
 
 | | Crystal Structure Analysis of (gp27-gp5)3 conjugated with Fe(III) protoporphyrin | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, Baseplate structural protein Gp27, Tail-associated lysozyme | | Authors: | Koshiyama, T, Yokoi, N, Ueno, T, Kanamaru, S, Nagano, S, Shiro, Y, Arisaka, F, Watanabe, Y. | | Deposit date: | 2007-07-28 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Molecular design of heteroprotein assemblies providing a bionanocup as a chemical reactor.
Small, 4, 2008
|
|
5WS7
 
 | | Crystal structure of human MTH1(G2K/C87A/C104S mutant) in complex with 2-oxo-dATP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-12-05 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
7WWX
 
 | | Crystal structure of Herbaspirillum huttiense L-arabinose 1-dehydrogenase (NAD bound form) | | Descriptor: | DI(HYDROXYETHYL)ETHER, NAD(P)-dependent dehydrogenase (Short-subunit alcohol dehydrogenase family), NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Matsubara, R, Yoshiwara, K, Watanabe, Y, Watanabe, S. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure of L-arabinose 1-dehydrogenase as a short-chain reductase/dehydrogenase protein.
Biochem.Biophys.Res.Commun., 604, 2022
|
|
2CXO
 
 | | Crystal structure of mouse AMF / E4P complex | | Descriptor: | D-4-PHOSPHOERYTHRONIC ACID, GLYCEROL, Glucose-6-phosphate isomerase | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
2CXT
 
 | | Crystal structure of mouse AMF / F6P complex | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, GLYCEROL, Glucose-6-phosphate isomerase | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
2CXN
 
 | | Crystal structure of mouse AMF / phosphate complex | | Descriptor: | GLYCEROL, Glucose-6-phosphate isomerase, PHOSPHATE ION | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
2CXU
 
 | | Crystal structure of mouse AMF / M6P complex | | Descriptor: | GLYCEROL, Glucose-6-phosphate isomerase, PHOSPHATE ION | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
6MUI
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-42 antibody | | Descriptor: | E1, E2, EEEV-42 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6MWV
 
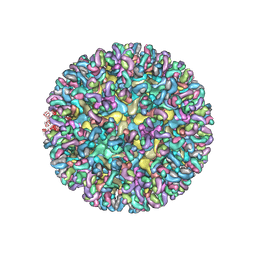 | | CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-58 Antibody | | Descriptor: | E1, E2, EEEV-58 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6MWC
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-5 antibody | | Descriptor: | E1, E2, EEEV-5 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-29 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
2CXP
 
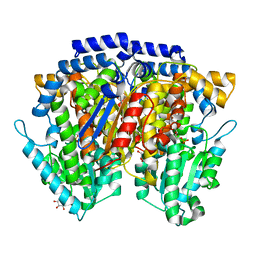 | | Crystal structure of mouse AMF / A5P complex | | Descriptor: | ARABINOSE-5-PHOSPHATE, GLYCEROL, Glucose-6-phosphate isomerase | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
2CVP
 
 | | Crystal structure of mouse AMF | | Descriptor: | ACETATE ION, GLYCEROL, Glucose-6-phosphate isomerase | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
2CXQ
 
 | | Crystal structure of mouse AMF / S6P complex | | Descriptor: | D-SORBITOL-6-PHOSPHATE, GLYCEROL, Glucose-6-phosphate isomerase | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
2E8J
 
 | |
2CXS
 
 | | Crystal structure of mouse AMF / F6P complex | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, GLYCEROL, Glucose-6-phosphate isomerase | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
2CXR
 
 | | Crystal structure of mouse AMF / 6PG complex | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, GLYCEROL, Glucose-6-phosphate isomerase | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
6MW9
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-3 antibody | | Descriptor: | E1, E2, EEEV-3 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-29 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
