5GQL
 
 | | Crystal structure of Wild Type Cypovirus Polyhedra | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
5GQJ
 
 | | Crystal structure of Cypovirus Polyhedra mutant with deletion of Ser193 and Ala194 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
5GQM
 
 | | Crystal structure of in cellulo Wild Type Cypovirus Polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
5GQK
 
 | | Crystal structure of Cypovirus Polyhedra mutant with deletion of Gly192-Ala194 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Polyhedrin | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
5GQN
 
 | | Crystal structure of in cellulo Cypovirus Polyhedra mutant with deletion of Gly192-Ala194 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Polyhedrin | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
5AXV
 
 | | Crystal Structure of Cypovirus Polyhedra R13K Mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Abe, S, Ijiri, H, Negishi, H, Yamanaka, H, Sasaki, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2015-08-01 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Design of Enzyme-Encapsulated Protein Containers by In Vivo Crystal Engineering
Adv. Mater. Weinheim, 27, 2015
|
|
5AXU
 
 | | Crystal Structure of Cypovirus Polyhedra R13A Mutant | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Abe, S, Ijiri, H, Negishi, H, Yamanaka, H, Sasaki, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2015-08-01 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Enzyme-Encapsulated Protein Containers by In Vivo Crystal Engineering
Adv. Mater. Weinheim, 27, 2015
|
|
6JEF
 
 | | Crystal structure of apo-R168C/L169C-Fr | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Abe, S, Ito, N, Maity, B, Lu, C, Lu, D, Ueno, T. | | Deposit date: | 2019-02-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Coordination design of cadmium ions at the 4-fold axis channel of the apo-ferritin cage.
Dalton Trans, 48, 2019
|
|
6JEE
 
 | | Crystal structure of apo-L161C/L165C-Fr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Abe, S, Ito, N, Maity, B, Lu, C, Lu, D, Ueno, T. | | Deposit date: | 2019-02-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Coordination design of cadmium ions at the 4-fold axis channel of the apo-ferritin cage.
Dalton Trans, 48, 2019
|
|
7E3G
 
 | | Crystal structure of Trypanosoma brucei cathepsin B R91C/T223C mutant in the living cell | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Abe, S, Pham, T.T, Negishi, H, Yamashita, K, Hirata, K, Ueno, T. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.86 Å) | | Cite: | Design of an In-Cell Protein Crystal for the Environmentally Responsive Construction of a Supramolecular Filament.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7E3E
 
 | | Crystal structure of Trypanosoma brucei cathepsin B R91C/T223C mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Abe, S, Pham, T.T, Negishi, H, Yamashita, K, Hirata, K, Ueno, T. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of an In-Cell Protein Crystal for the Environmentally Responsive Construction of a Supramolecular Filament.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7E3F
 
 | | Crystal structure of Trypanosoma brucei cathepsin B Y217C/S275C mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Abe, S, Pham, T.T, Negishi, H, Yamashita, K, Hirata, K, Ueno, T. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Design of an In-Cell Protein Crystal for the Environmentally Responsive Construction of a Supramolecular Filament.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
3AZ4
 
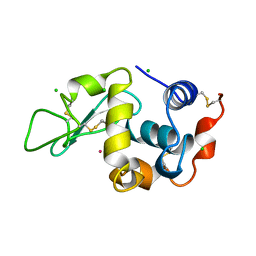 | | Crystal structure of Co/O-HEWL | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Lysozyme C | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3AZ5
 
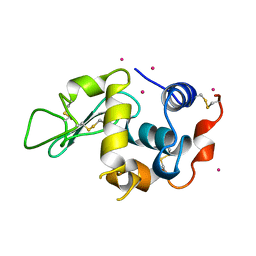 | | Crystal structure of Pt/O-HEWL | | Descriptor: | Lysozyme C, PLATINUM (II) ION | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3AZ6
 
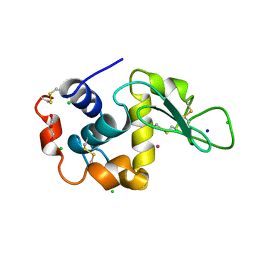 | | Crystal structure of Co/T-HEWL | | Descriptor: | CHLORIDE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3AZ7
 
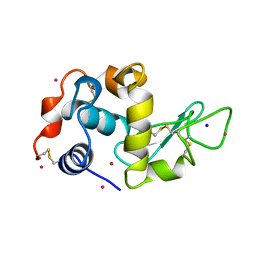 | | Crystal structure of Pt/T-HEWL | | Descriptor: | Lysozyme C, PLATINUM (II) ION, SODIUM ION | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
5YHA
 
 | | Crystal structure of Pd(allyl)/Wild Type Polyhedra | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PALLADIUM ION, ... | | Authors: | Abe, S, Atsumi, K, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of in cell protein crystals containing organometallic complexes.
Phys Chem Chem Phys, 20, 2018
|
|
5YHB
 
 | | Crystal structure of Pd(allyl)/polyhedra mutant with deletion of Gly192-Ala194 | | Descriptor: | PALLADIUM ION, Palladium(II) allyl complex, Polyhedrin | | Authors: | Abe, S, Atsumi, K, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of in cell protein crystals containing organometallic complexes.
Phys Chem Chem Phys, 20, 2018
|
|
6IGJ
 
 | | Crystal structure of FT condition 4 | | Descriptor: | MAGNESIUM ION, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
6IGG
 
 | | Crystal structure of FT condition 1 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
6IGH
 
 | | Crystal structure of FT condition3 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
6IGI
 
 | | Crystal structure of FT condition 2 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
2D2E
 
 | | Crystal structure of atypical cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, GLYCEROL, SufC protein | | Authors: | Watanabe, S, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-09-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Atypical Cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8.
J.Mol.Biol., 353, 2005
|
|
3VYT
 
 | | Crystal structure of the HypC-HypD-HypE complex (form I inward) | | Descriptor: | CHLORIDE ION, Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
3VYR
 
 | | Crystal structure of the HypC-HypD complex | | Descriptor: | CITRIC ACID, Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
