1Y9Z
 
 | |
1HZE
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF RIBOFLAVIN SYNTHASE FROM E. COLI | | Descriptor: | RIBOFLAVIN, RIBOFLAVIN SYNTHASE ALPHA CHAIN | | Authors: | Truffault, V, Coles, M, Diercks, T, Abelmann, K, Eberhardt, S, Luettgen, H, Bacher, A, Kessler, H. | | Deposit date: | 2001-01-24 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of riboflavin synthase.
J.Mol.Biol., 309, 2001
|
|
1I18
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF RIBOFLAVIN SYNTHASE FROM E. COLI | | Descriptor: | RIBOFLAVIN, RIBOFLAVIN SYNTHASE ALPHA CHAIN | | Authors: | Truffault, V, Coles, M, Diercks, T, Abelmann, K, Eberhardt, S, Luettgen, H, Bacher, A, Kessler, H. | | Deposit date: | 2001-01-31 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of riboflavin synthase.
J.Mol.Biol., 309, 2001
|
|
1J1T
 
 | | Alginate lyase from Alteromonas sp.272 | | Descriptor: | Alginate Lyase, CALCIUM ION, SULFATE ION | | Authors: | Motoshima, H, Iwatomo, Y, Watanabe, K, Oda, T, Muramatsu, T. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Alginate Lyase from Alteromonas sp.272
To be published
|
|
6J6C
 
 | |
6J5N
 
 | |
6J5M
 
 | |
6J5Q
 
 | |
6J6E
 
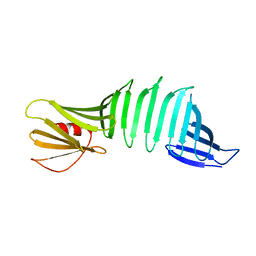 | |
6J5O
 
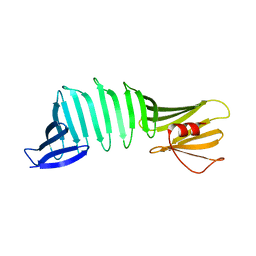 | |
6J6B
 
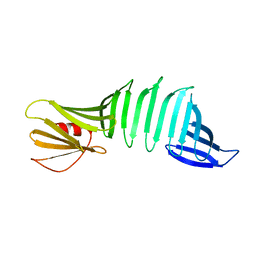 | |
6J5P
 
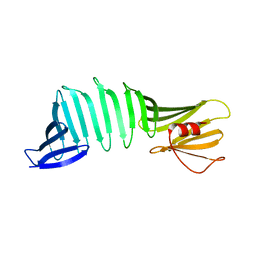 | |
6J5R
 
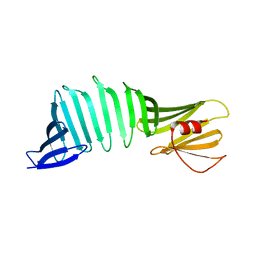 | |
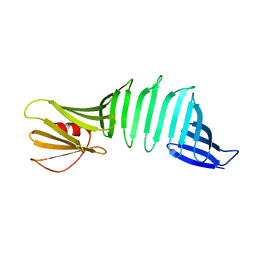
6J6D
 
 | |
2EK8
 
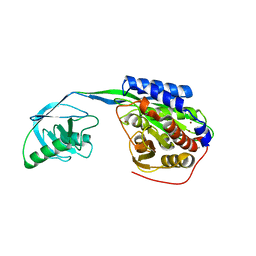 | |
2F38
 
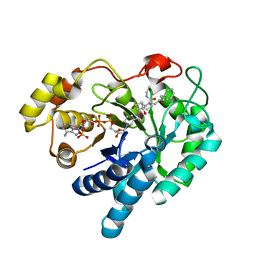 | | Crystal structure of prostaglandin F synathase containing bimatoprost | | Descriptor: | (5Z)-7-{(1R,2R,3R,5S)-3,5-DIHYDROXY-2-[(1E,3S)-3-HYDROXY-5-PHENYLPENT-1-ENYL]CYCLOPENTYL}-N-ETHYLHEPT-5-ENAMIDE, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Komoto, J, Yamada, T, Watanabe, K, Woodward, D.F, Takusagawa, F. | | Deposit date: | 2005-11-18 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prostaglandin F2alpha formation from prostaglandin H2 by prostaglandin F synthase (PGFS): crystal structure of PGFS containing bimatoprost.
Biochemistry, 45, 2006
|
|
2HDN
 
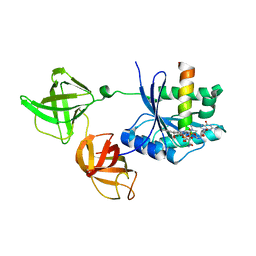 | | Trypsin-modified Elongation Factor Tu in complex with tetracycline at 2.8 Angstrom resolution | | Descriptor: | Elongation factor EF-Tu, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mui, S, Heffron, S.E, Aorora, A, Abel, K, Bergmann, E, Jurnak, F. | | Deposit date: | 2006-06-20 | | Release date: | 2006-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular complementarity between tetracycline and the GTPase active site of elongation factor Tu.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2HCJ
 
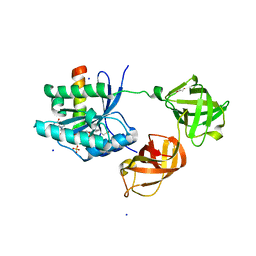 | | Trypsin-modified Elongation Factor Tu in complex with tetracycline | | Descriptor: | GLYOXYLIC ACID, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mui, S, Heffron, S.E, Aorora, A, Abel, K, Bergmann, E, Jurnak, F. | | Deposit date: | 2006-06-16 | | Release date: | 2006-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Molecular complementarity between tetracycline and the GTPase active site of elongation factor Tu.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6IDC
 
 | |
6IYS
 
 | |
6K0U
 
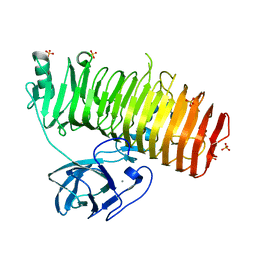 | | Catalytic domain of GH87 alpha-1,3-glucanase D1068A in complex with tetrasaccharides | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0M
 
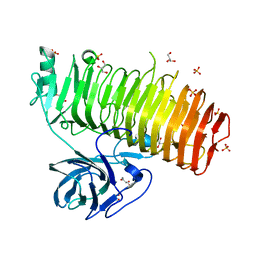 | | Catalytic domain of GH87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11 | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, GLYCEROL, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0Q
 
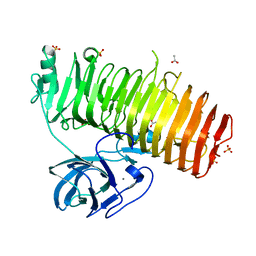 | | Catalytic domain of GH87 alpha-1,3-glucanase D1068A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.564 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0P
 
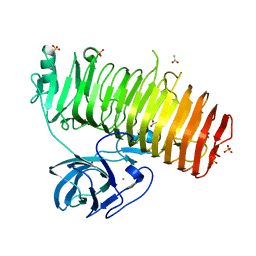 | | Catalytic domain of GH87 alpha-1,3-glucanase D1045A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0N
 
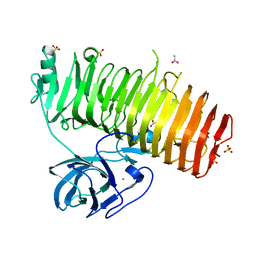 | | Catalytic domain of GH87 alpha-1,3-glucanase in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
