5L9Q
 
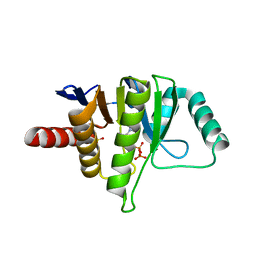 | | OCEANOBACILLUS IHEYENSIS MACRODOMAIN WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MACROD-TYPE MACRODOMAIN, SULFATE ION | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-10 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
4FCT
 
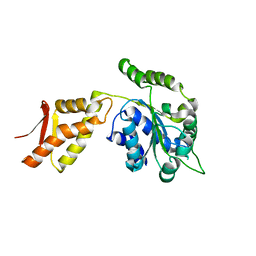 | |
4GDP
 
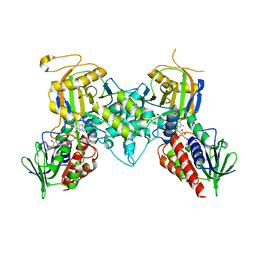 | | Yeast polyamine oxidase FMS1, N195A mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1, TETRAETHYLENE GLYCOL | | Authors: | Taylor, A.B, Adachi, M.S, Hart, P.J, Fitzpatrick, P.F. | | Deposit date: | 2012-08-01 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9998 Å) | | Cite: | Mechanistic and Structural Analyses of the Roles of Active Site Residues in Yeast Polyamine Oxidase Fms1: Characterization of the N195A and D94N Enzymes.
Biochemistry, 51, 2012
|
|
5L9K
 
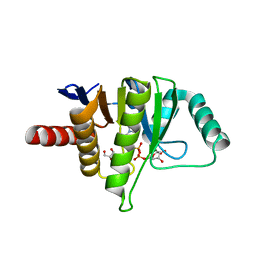 | | OCEANOBACILLUS IHEYENSIS MACRODOMAIN WITH ADPR | | Descriptor: | GLYCEROL, MACROD-TYPE MACRODOMAIN, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-10 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
4FUP
 
 | |
4ECH
 
 | | Yeast Polyamine Oxidase FMS1, H67Q Mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1 | | Authors: | Taylor, A.B, Adachi, M.S, Hart, P.J, Fitzpatrick, P.F. | | Deposit date: | 2012-03-26 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic and structural analyses of the role of his67 in the yeast polyamine oxidase fms1.
Biochemistry, 51, 2012
|
|
5LCC
 
 | | Oceanobacillus iheyensis macrodomain mutant D40A | | Descriptor: | MACROD-TYPE MACRODOMAIN | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
4FDM
 
 | | Crystallization and 3D structure elucidation of thermostable L2 lipase from thermophilic locally isolated Bacillus sp. L2. | | Descriptor: | CALCIUM ION, Thermostable lipase, ZINC ION | | Authors: | Rahman, R.N.Z.R.A, Shariff, F.M, Salleh, A.B, Basri, M.B. | | Deposit date: | 2012-05-29 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 3D Structure Elucidation of Thermostable L2 Lipase from Thermophilic Bacillus sp. L2.
Int.J.Mol.Sci., 13, 2012
|
|
4FD2
 
 | | Crystal structure of the C-terminal domain of ClpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Biter, A.B, Lee, S, Sung, N, Tsai, F.T.F. | | Deposit date: | 2012-05-25 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for intersubunit signaling in a protein disaggregating machine.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HPM
 
 | | PCGF1 Ub fold (RAWUL)/BCORL1 PUFD Complex | | Descriptor: | BCL-6 corepressor-like protein 1, PHOSPHATE ION, Polycomb group RING finger protein 1 | | Authors: | Junco, S.E, Wang, R, Gaipa, J, Taylor, A.B, Gearhart, M.D, Bardwell, V.J, Hart, P.J, Kim, C.A. | | Deposit date: | 2012-10-24 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the Polycomb Group Protein PCGF1 in Complex with BCOR Reveals Basis for Binding Selectivity of PCGF Homologs.
Structure, 21, 2013
|
|
4FUO
 
 | |
4FUN
 
 | |
4HPL
 
 | | PCGF1 Ub fold (RAWUL)/BCOR PUFD Complex | | Descriptor: | BCL-6 corepressor, Polycomb group RING finger protein 1 | | Authors: | Junco, S.E, Wang, R, Gaipa, J, Taylor, A.B, Gearhart, M.D, Bardwell, V.J, Hart, P.J, Kim, C.A. | | Deposit date: | 2012-10-24 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Polycomb Group Protein PCGF1 in Complex with BCOR Reveals Basis for Binding Selectivity of PCGF Homologs.
Structure, 21, 2013
|
|
1G6O
 
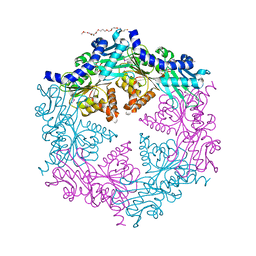 | | CRYSTAL STRUCTURE OF THE HELICOBACTER PYLORI ATPASE, HP0525, IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CAG-ALPHA, DI(HYDROXYETHYL)ETHER | | Authors: | Yeo, H.J, Savvides, S.N, Herr, A.B, Lanka, E, Waksman, G, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-11-07 | | Release date: | 2001-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system.
Mol.Cell, 6, 2000
|
|
1HR6
 
 | | Yeast Mitochondrial Processing Peptidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1QZQ
 
 | | human Tyrosyl DNA phosphodiesterase | | Descriptor: | tyrosyl-DNA phosphodiesterase 1 | | Authors: | Raymond, A.C, Rideout, M.C, Staker, B, Hjerrild, K, Burgin Jr, A.B. | | Deposit date: | 2003-09-17 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Analysis of Human Tyrosyl-DNA Phosphodiesterase I Catalytic Residues.
J.Mol.Biol., 338, 2004
|
|
1QVS
 
 | | Crystal Structure of Haemophilus influenzae H9A mutant Holo Ferric ion-Binding Protein A | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein, PHOSPHATE ION | | Authors: | Shouldice, S.R, Skene, R.J, Dougan, D.R, McRee, D.E, Tari, L.W, Schryvers, A.B. | | Deposit date: | 2003-08-28 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Presence of ferric hydroxide clusters in mutants of Haemophilus influenzae ferric ion-binding protein A
Biochemistry, 42, 2003
|
|
1QW0
 
 | | Crystal Structure of Haemophilus influenzae N175L mutant Holo Ferric ion-Binding Protein A | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein, PHOSPHATE ION | | Authors: | Shouldice, S.R, Skene, R.J, Dougan, D.R, McRee, D.E, Tari, L.W, Schryvers, A.B. | | Deposit date: | 2003-08-29 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Presence of ferric hydroxide clusters in mutants of Haemophilus influenzae ferric ion-binding protein A
Biochemistry, 42, 2003
|
|
1HR8
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Cytochrome C Oxidase IV Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYTOCHROME C OXIDASE POLYPEPTIDE IV, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1HZS
 
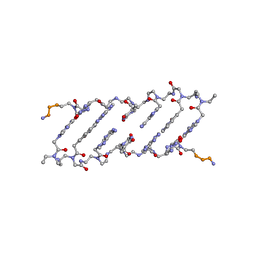 | | Crystal structure of a peptide nucleic acid duplex (BT-PNA) containing a bicyclic analogue of thymine | | Descriptor: | PEPTIDE NUCLEIC ACID | | Authors: | Eldrup, A.B, Nielsen, B.B, Haaima, G, Rasmussen, H, Kastrup, J.S, Christensen, C, Nielsen, P.E. | | Deposit date: | 2001-01-26 | | Release date: | 2001-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 1,8-Naphthyridin-2(1H)-ones. Novel Bicyclic and Tricyclic Analogues of Thymine in Peptide Nucleic Acids (PNAs)
Eur.J.Org.Chem., 9, 2001
|
|
1I1G
 
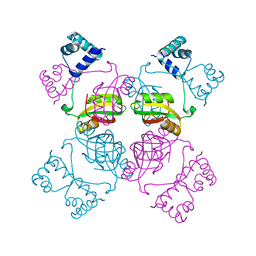 | | CRYSTAL STRUCTURE OF THE LRP-LIKE TRANSCRIPTIONAL REGULATOR FROM THE ARCHAEON PYROCOCCUS FURIOSUS | | Descriptor: | TRANSCRIPTIONAL REGULATOR LRPA | | Authors: | Leonard, P.M, Smits, S.H.J, Sedelnikova, S.E, Brinkman, A.B, de Vos, W.M, van der Oost, J, Rice, D.W, Rafferty, J.B. | | Deposit date: | 2001-02-01 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus.
EMBO J., 20, 2001
|
|
1OIF
 
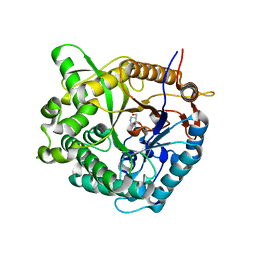 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, BETA-GLUCOSIDASE | | Authors: | Gloster, T, Zechel, D, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-06-16 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
1OIM
 
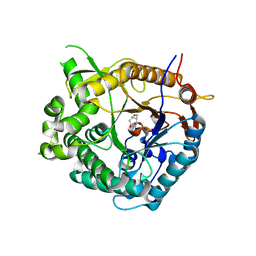 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | 1-DEOXYNOJIRIMYCIN, BETA-GLUCOSIDASE A | | Authors: | Gloster, T, Zechel, D.L, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
1IC0
 
 | | RED COPPER PROTEIN NITROSOCYANIN FROM NITROSOMONAS EUROPAEA | | Descriptor: | COPPER (II) ION, Nitrosocyanin | | Authors: | Lieberman, R.L, Arciero, D.M, Hooper, A.B, Rosenzweig, A.C. | | Deposit date: | 2001-03-29 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a novel red copper protein from Nitrosomonas europaea.
Biochemistry, 40, 2001
|
|
2L21
 
 | | chicken IGF2R domain 11 | | Descriptor: | Cation-independent mannose-6-phosphate receptor | | Authors: | Williams, C, Hoppe, H, Strickland, M, Frago, S, Ellis, R.Z, Wattana-Amorn, P, Prince, S.N, Zaccheo, O.J, Forbes, B, Jones, E.Y, Rezgui, D.Z, Crump, M.P, Hassan, A.B. | | Deposit date: | 2010-08-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | CD loop dependency of the IGF2: M6P/IGF2 receptor binding interaction predates imprinting
To be Published
|
|
