1G7O
 
 | | NMR SOLUTION STRUCTURE OF REDUCED E. COLI GLUTAREDOXIN 2 | | 分子名称: | GLUTAREDOXIN 2 | | 著者 | Xia, B, Vlamis-Gardikas, A, Holmgren, A, Wright, P.E, Dyson, H.J. | | 登録日 | 2000-11-10 | | 公開日 | 2001-07-20 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Escherichia coli glutaredoxin-2 shows similarity to mammalian glutathione-S-transferases.
J.Mol.Biol., 310, 2001
|
|
2B9K
 
 | |
2LIZ
 
 | |
6K5X
 
 | |
2HLU
 
 | |
2HLT
 
 | |
2LTU
 
 | |
7F7N
 
 | |
5ZUX
 
 | |
5ZUZ
 
 | |
2N9X
 
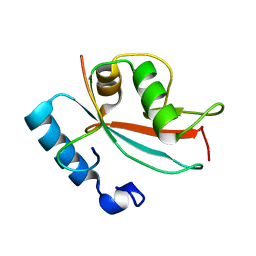 | | LC3 FUNDC1 complex structure | | 分子名称: | FUN14 domain-containing protein 1, Microtubule-associated proteins 1A/1B light chain 3B | | 著者 | Xia, B, Kuang, Y. | | 登録日 | 2015-12-14 | | 公開日 | 2016-12-14 | | 最終更新日 | 2017-06-07 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the phosphorylation of FUNDC1 LIR as a molecular switch of mitophagy.
Autophagy, 12, 2016
|
|
6JYK
 
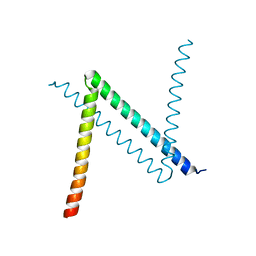 | |
6K2J
 
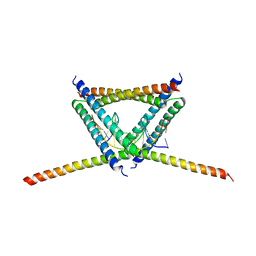 | |
2K7X
 
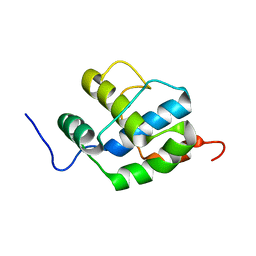 | |
2FHM
 
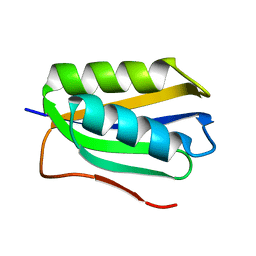 | |
6E4H
 
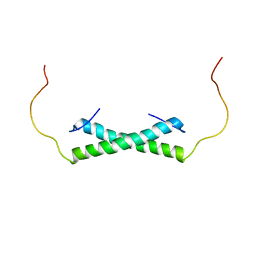 | | Solution NMR Structure of the Colied-coil PALB2 Homodimer | | 分子名称: | Partner and localizer of BRCA2 | | 著者 | Song, F, Li, M, Liu, G, Swapna, G.V.T, Xia, B, Bunting, S.F, Montelione, G.T. | | 登録日 | 2018-07-17 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Antiparallel Coiled-Coil Interactions Mediate the Homodimerization of the DNA Damage-Repair Protein PALB2.
Biochemistry, 57, 2018
|
|
3BR8
 
 | |
3IWM
 
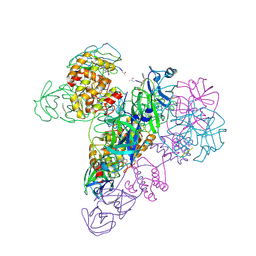 | | The octameric SARS-CoV main protease | | 分子名称: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Zhong, N, Zhang, S, Xue, F, Lou, Z, Rao, Z, Xia, B. | | 登録日 | 2009-09-02 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Three-dimensional domain swapping as a mechanism to lock the active conformation in a super-active octamer of SARS-CoV main protease
Protein Cell, 1, 2010
|
|
3EBN
 
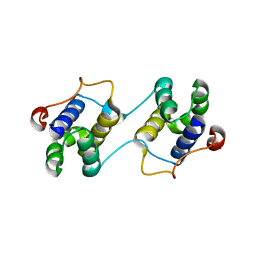 | | A Special Dimerization of SARS-CoV Main Protease C-Terminal Domain Due to Domain-swapping | | 分子名称: | Replicase polyprotein 1ab | | 著者 | Zhong, N, Zhang, S, Xue, F, Kang, X, Lou, Z, Xia, B. | | 登録日 | 2008-08-28 | | 公開日 | 2009-05-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | C-terminal domain of SARS-CoV main protease can form a 3D domain-swapped dimer
PROTEIN SCI., 18, 2009
|
|
2K4K
 
 | | Solution structure of GSP13 from Bacillus subtilis | | 分子名称: | General stress protein 13 | | 著者 | Yu, W, Yu, B, Hu, J, Jin, C, Xia, B. | | 登録日 | 2008-06-13 | | 公開日 | 2009-05-12 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of GSP13 from Bacillus subtilis exhibits an S1 domain related to cold shock proteins.
J.Biomol.Nmr, 43, 2009
|
|
1Z6H
 
 | |
2M5H
 
 | |
2MXF
 
 | |
2MXE
 
 | |
1Z7R
 
 | | Solution Structure of reduced glutaredoxin C1 from Populus tremula x tremuloides | | 分子名称: | glutaredoxin | | 著者 | Feng, Y, Zhong, N, Rouhier, N, Jacquot, J.P, Xia, B. | | 登録日 | 2005-03-26 | | 公開日 | 2006-03-28 | | 最終更新日 | 2020-02-26 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insight into Poplar Glutaredoxin C1 with a Bridging Iron-Sulfur Cluster at the Active Site
Biochemistry, 45, 2006
|
|
