4RUW
 
 | |
6VC5
 
 | |
6UAG
 
 | |
6UG4
 
 | |
6UHS
 
 | |
6UHH
 
 | | Crystal Structure of Human RYR Receptor 3 ( 848-1055) in Complex with ATP | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wu, R, Kim, Y, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2019-09-27 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.138 Å) | | 主引用文献 | Crystal Structure of Human RYR Receptor 3 ( 848-1055) in Complex with ATP
To Be Published
|
|
6UHA
 
 | |
6UHB
 
 | |
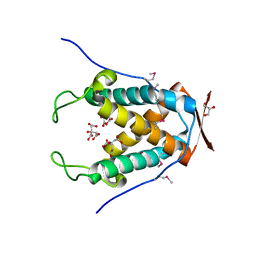
6UAM
 
 | |
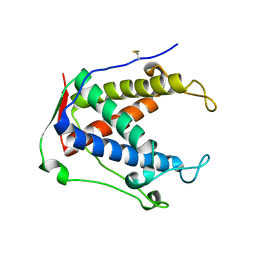
6UHE
 
 | |
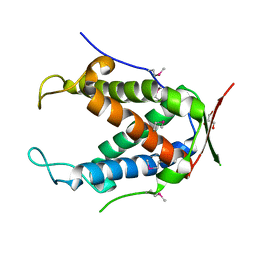
6UHI
 
 | |
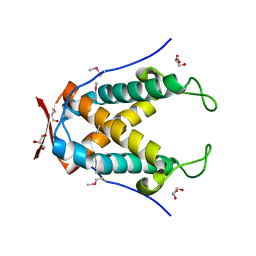
6UG5
 
 | |
1RLJ
 
 | | Structural Genomics, a Flavoprotein NrdI from Bacillus subtilis | | 分子名称: | FLAVIN MONONUCLEOTIDE, IODIDE ION, NrdI protein | | 著者 | Wu, R, Zhang, R, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-11-25 | | 公開日 | 2004-07-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 1.5A crystal structure of a thioredoxin-like protein NrdI from Bacillus subtilis
To be Published
|
|
1XFK
 
 | |
4RBN
 
 | | The crystal structure of Nitrosomonas europaea sucrose synthase: Insights into the evolutionary origin of sucrose metabolism in prokaryotes | | 分子名称: | Sucrose synthase:Glycosyl transferases group 1 | | 著者 | Wu, R, Asencion Diez, M.D, Figueroa, C.M, Machtey, M, Iglesias, A.A, Ballicora, M.A, Liu, D. | | 登録日 | 2014-09-12 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | The Crystal Structure of Nitrosomonas europaea Sucrose Synthase Reveals Critical Conformational Changes and Insights into Sucrose Metabolism in Prokaryotes.
J.Bacteriol., 197, 2015
|
|
4RJ0
 
 | |
4RIZ
 
 | |
4RIT
 
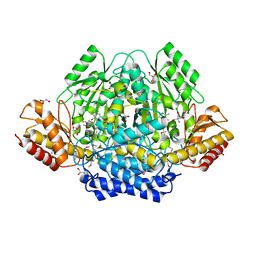 | | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-10-07 | | 公開日 | 2014-10-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745
To be Published
|
|
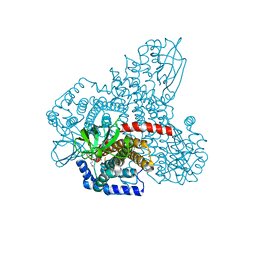
4RM7
 
 | |
4RT5
 
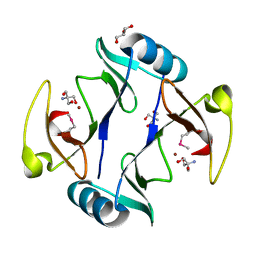 | | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase protein from planctomyces limnophilus dsm 3776 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | 著者 | Wu, R, Bearden, J, Kim, Y, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-13 | | 公開日 | 2014-12-03 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase protein from Planctomyces limnophilus dsm 3776
TO BE PUBLISHED
|
|
4RU0
 
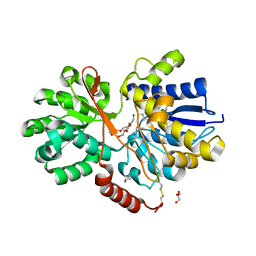 | | The crystal structure of abc transporter permease from pseudomonas fluorescens group | | 分子名称: | 3,6,9,12,15-PENTAOXAHEPTADECANE, GLYCEROL, Putative branched-chain amino acid ABC transporter, ... | | 著者 | Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-17 | | 公開日 | 2014-11-26 | | 実験手法 | X-RAY DIFFRACTION (2.442 Å) | | 主引用文献 | The crystal structure of abc transporter permease from pseudomonas fluorescens group
To be Published
|
|
4RM1
 
 | |
4RLG
 
 | |
6VC6
 
 | | 2.1 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Mn2+ | | 分子名称: | 6-O-phosphono-alpha-D-glucopyranose, 6-phospho-alpha-glucosidase, GLYCEROL, ... | | 著者 | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | 登録日 | 2019-12-20 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.133 Å) | | 主引用文献 | 2.1 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Mn2+
To Be Published
|
|
6WBT
 
 | | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate | | 分子名称: | 1,2-ETHANEDIOL, 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, ... | | 著者 | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | 登録日 | 2020-03-27 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate
To Be Published
|
|
