3PYW
 
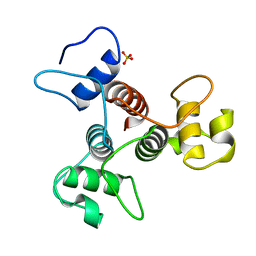 | | The structure of the SLH domain from B. anthracis surface array protein at 1.8A | | 分子名称: | S-layer protein sap, SULFATE ION | | 著者 | Zhang, R, Wilton, R, Kern, J, Joachimiak, A, Schneewind, O, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-12-13 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of Surface Layer Homology (SLH) Domains from Bacillus anthracis Surface Array Protein.
J.Biol.Chem., 286, 2011
|
|
3N04
 
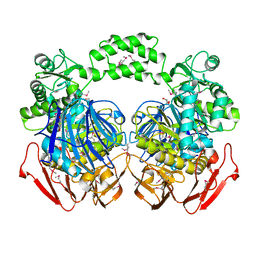 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (FAMILY 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 | | 分子名称: | GLYCEROL, alpha-glucosidase | | 著者 | Tan, K, Tesar, C, Freeman, L, Wilton, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-05-13 | | 公開日 | 2010-06-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (FAMILY 31) FROM RUMINOCOCCUS OBEUM ATCC 29174
Faseb J., 24, 2010
|
|
3NUK
 
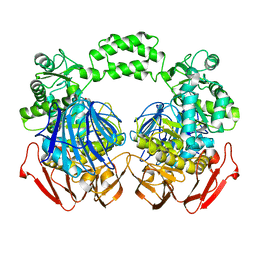 | | THE CRYSTAL STRUCTURE OF THE W169Y mutant of ALPHA-GLUCOSIDASE (FAMILY 31) from RUMINOCOCCUS OBEUM ATCC 29174 | | 分子名称: | ALPHA-GLUCOSIDASE, GLYCEROL | | 著者 | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-07-07 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.055 Å) | | 主引用文献 | THE CRYSTAL STRUCTURE OF THE W169Y mutant of ALPHA-GLUCOSIDASE (FAMILY 31) from RUMINOCOCCUS OBEUM ATCC 29174
TO BE PUBLISHED
|
|
3NSX
 
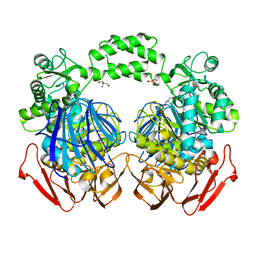 | | The crystal structure of the The crystal structure of the D420A mutant of the alpha-glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-glucosidase | | 著者 | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-07-02 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.569 Å) | | 主引用文献 | The crystal structure of the The crystal structure of the D420A mutant of the alpha-glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174
To be Published
|
|
3POC
 
 | | The crystal structure of the D307A mutant of alpha-Glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with acarbose | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLYCEROL, ... | | 著者 | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-11-22 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | The crystal structure of the D307A mutant of alpha-Glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with acarbose
To be Published
|
|
3UMT
 
 | |
3USB
 
 | | Crystal Structure of Bacillus anthracis Inosine Monophosphate Dehydrogenase in the complex with IMP | | 分子名称: | CHLORIDE ION, GLYCEROL, INOSINIC ACID, ... | | 著者 | Kim, Y, Zhang, R, Wu, R, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-11-23 | | 公開日 | 2011-12-07 | | 最終更新日 | 2019-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
3TSD
 
 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames complexed with XMP | | 分子名称: | D(-)-TARTARIC ACID, Inosine-5'-monophosphate dehydrogenase, SULFATE ION, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-13 | | 公開日 | 2011-10-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.653 Å) | | 主引用文献 | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
3TSB
 
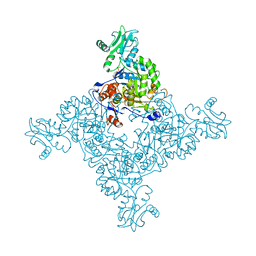 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames | | 分子名称: | Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION | | 著者 | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-12 | | 公開日 | 2011-10-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.595 Å) | | 主引用文献 | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
6NEX
 
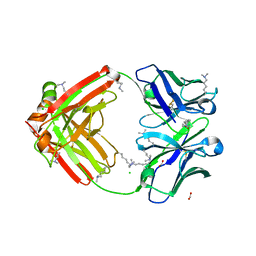 | | Fab fragment of anti-cocaine antibody h2E2 | | 分子名称: | ACETATE ION, Anitgen binding fragment light chain, Antigen binding fragment heavy chain, ... | | 著者 | Pokkuluri, P.R, Tan, K. | | 登録日 | 2018-12-18 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural analysis of free and liganded forms of the Fab fragment of a high-affinity anti-cocaine antibody, h2E2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6NFN
 
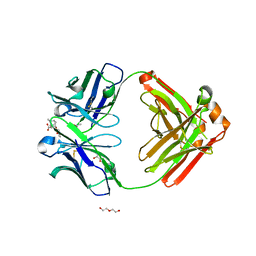 | | Fab fragment of anti-cocaine antibody h2E2 bound to benzoylecgonine | | 分子名称: | 3-(BENZOYLOXY)-8-METHYL-8-AZABICYCLO[3.2.1]OCTANE-2-CARBOXYLIC ACID, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Pokkuluri, P.R, Tan, K. | | 登録日 | 2018-12-20 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Structural analysis of free and liganded forms of the Fab fragment of a high-affinity anti-cocaine antibody, h2E2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
3B47
 
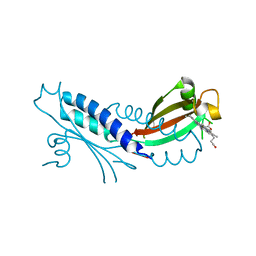 | | Periplasmic sensor domain of chemotaxis protein GSU0582 | | 分子名称: | Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Pokkuluri, P.R, Schiffer, M. | | 登録日 | 2007-10-23 | | 公開日 | 2008-04-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures and solution properties of two novel periplasmic sensor domains with c-type heme from chemotaxis proteins of Geobacter sulfurreducens: implications for signal transduction.
J.Mol.Biol., 377, 2008
|
|
3B42
 
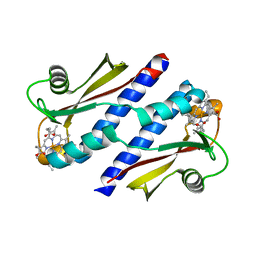 | | Periplasmic sensor domain of chemotaxis protein GSU0935 | | 分子名称: | Methyl-accepting chemotaxis protein, putative, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Pokkuluri, P.R, Schiffer, M. | | 登録日 | 2007-10-23 | | 公開日 | 2008-04-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures and solution properties of two novel periplasmic sensor domains with c-type heme from chemotaxis proteins of Geobacter sulfurreducens: implications for signal transduction.
J.Mol.Biol., 377, 2008
|
|
4JGP
 
 | | The crystal structure of sporulation kinase D sensor domain from Bacillus subtilis subsp in complex with pyruvate at 2.0A resolution | | 分子名称: | PYRUVIC ACID, Sporulation kinase D | | 著者 | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-03-01 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4JGQ
 
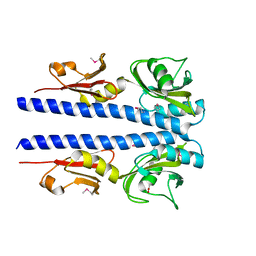 | | The crystal structure of sporulation kinase D mutant sensor domain, r131a, from Bacillus subtilis subsp in co-crystallization with pyruvate | | 分子名称: | ACETIC ACID, Sporulation kinase D | | 著者 | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-03-01 | | 公開日 | 2013-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4JGO
 
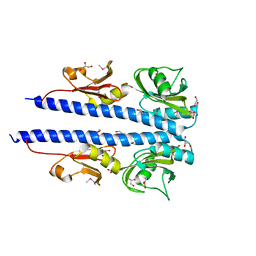 | | The crystal structure of sporulation kinase d sensor domain from Bacillus subtilis subsp. | | 分子名称: | GLYCEROL, PYRUVIC ACID, Sporulation kinase D | | 著者 | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-03-01 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4JGR
 
 | | The crystal structure of sporulation kinase D mutant sensor domain, R131A, from Bacillus subtilis subsp at 2.4A resolution | | 分子名称: | ACETIC ACID, GLYCEROL, Sporulation kinase D | | 著者 | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-03-01 | | 公開日 | 2013-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4K08
 
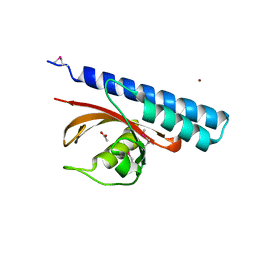 | | Periplasmic sensor domain of chemotaxis protein, Adeh_3718 | | 分子名称: | ACETATE ION, Methyl-accepting chemotaxis sensory transducer, ZINC ION | | 著者 | Pokkuluri, P.R, Mack, J.C, Bearden, J, Rakowski, E, Schiffer, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-04-03 | | 公開日 | 2013-07-17 | | 最終更新日 | 2015-04-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Analysis of periplasmic sensor domains from Anaeromyxobacter dehalogenans 2CP-C: structure of one sensor domain from a histidine kinase and another from a chemotaxis protein.
Microbiologyopen, 2, 2013
|
|
4K0D
 
 | | Periplasmic sensor domain of sensor histidine kinase, Adeh_2942 | | 分子名称: | ACETATE ION, CHLORIDE ION, Periplasmic sensor hybrid histidine kinase, ... | | 著者 | Pokkuluri, P.R, Mack, J.C, Bearden, J, Rakowski, E, Schiffer, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-04-03 | | 公開日 | 2013-06-12 | | 最終更新日 | 2015-04-15 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | Analysis of periplasmic sensor domains from Anaeromyxobacter dehalogenans 2CP-C: structure of one sensor domain from a histidine kinase and another from a chemotaxis protein.
Microbiologyopen, 2, 2013
|
|
3M6D
 
 | | The crystal structure of the d307a mutant of glycoside Hydrolase (family 31) from ruminococcus obeum atcc 29174 | | 分子名称: | Uncharacterized protein | | 著者 | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-03-15 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Novel alpha-glucosidase from human gut microbiome: substrate specificities and their switch.
Faseb J., 24, 2010
|
|
3MKK
 
 | | The crystal structure of the D307A mutant of glycoside HYDROLASE (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with isomaltose | | 分子名称: | alpha-D-glucopyranose, alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose, alpha-glucosidase GH31 family, ... | | 著者 | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-04-15 | | 公開日 | 2010-06-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | The Crystal Structures Of The Glycoside Hydrolase (Family 31) From Ruminococcus Obeum Atcc 29174
Faseb J., 24, 2010
|
|
3M46
 
 | | The crystal structure of the D73A mutant of glycoside HYDROLASE (FAMILY 31) from Ruminococcus obeum ATCC 29174 | | 分子名称: | GLYCEROL, Uncharacterized protein | | 著者 | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-03-10 | | 公開日 | 2010-03-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Novel alpha-glucosidase from human gut microbiome: substrate specificities and their switch
Faseb J., 24, 2010
|
|
