7ZC8
 
 | |
1F0K
 
 | | THE 1.9 ANGSTROM CRYSTAL STRUCTURE OF E. COLI MURG | | 分子名称: | SULFATE ION, UDP-N-ACETYLGLUCOSAMINE-N-ACETYLMURAMYL-(PENTAPEPTIDE) PYROPHOSPHORYL-UNDECAPRENOL N-ACETYLGLUCOSAMINE TRANSFERASE | | 著者 | Ha, S, Walker, D, Shi, Y, Walker, S. | | 登録日 | 2000-05-16 | | 公開日 | 2000-07-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The 1.9 A crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis.
Protein Sci., 9, 2000
|
|
1NLM
 
 | | CRYSTAL STRUCTURE OF MURG:GLCNAC COMPLEX | | 分子名称: | GLYCEROL, UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | 著者 | Hu, Y, Chen, L, Ha, S, Gross, B, Falcone, B, Walker, D, Mokhtarzadeh, M, Walker, S. | | 登録日 | 2003-01-07 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4ZHO
 
 | | The crystal structure of Arabidopsis ferredoxin 2 with 2Fe-2S cluster | | 分子名称: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-2, ... | | 著者 | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | 登録日 | 2015-04-26 | | 公開日 | 2016-08-31 | | 最終更新日 | 2017-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
4ZHP
 
 | | The crystal structure of Potato ferredoxin I with 2Fe-2S cluster | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, Potato Ferredoxin I | | 著者 | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | 登録日 | 2015-04-26 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
4ZGV
 
 | | The Crystal Structure of the Ferredoxin Receptor FusA from Pectobacterium atrosepticum SCRI1043 | | 分子名称: | Ferredoxin receptor, LAURYL DIMETHYLAMINE-N-OXIDE, octyl beta-D-glucopyranoside | | 著者 | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | 登録日 | 2015-04-24 | | 公開日 | 2016-08-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
1E44
 
 | | ribonuclease domain of colicin E3 in complex with its immunity protein | | 分子名称: | 1,2-ETHANEDIOL, COLICIN E3, IMMUNITY PROTEIN | | 著者 | Carr, S, Walker, D, James, R, Kleanthous, C, Hemmings, A.M. | | 登録日 | 2000-06-28 | | 公開日 | 2001-06-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Inhibition of a Ribosome Inactivating Ribonuclease: The Crystal Structure of the Cytotoxic Domain of Colicin E3 in Complex with its Immunity Protein
Structure, 8, 2000
|
|
4QAY
 
 | |
4RTD
 
 | | Escherichia coli alpha-2-macroglobulin activated by porcine elastase | | 分子名称: | Uncharacterized lipoprotein YfhM | | 著者 | Fyfe, C.D, Grinter, R, Roszak, A.W, Josts, I, Cogdell, R.J, Walker, D. | | 登録日 | 2014-11-14 | | 公開日 | 2015-07-15 | | 最終更新日 | 2015-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | Structure of protease-cleaved Escherichia coli alpha-2-macroglobulin reveals a putative mechanism of conformational activation for protease entrapment.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4FZL
 
 | | High resolution structure of truncated bacteriocin syringacin M from Pseudomonas syringae pv. tomato DC3000 | | 分子名称: | 1,2-ETHANEDIOL, Bacteriocin, CALCIUM ION, ... | | 著者 | Roszak, A.W, Grinter, R, Cogdell, J.R, Walker, D. | | 登録日 | 2012-07-06 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | The Crystal Structure of the Lipid II-degrading Bacteriocin Syringacin M Suggests Unexpected Evolutionary Relationships between Colicin M-like Bacteriocins.
J.Biol.Chem., 287, 2012
|
|
4FZN
 
 | |
4FZM
 
 | | Crystal structure of the bacteriocin syringacin M from Pseudomonas syringae pv. tomato DC3000 | | 分子名称: | 1,2-ETHANEDIOL, Bacteriocin, CALCIUM ION | | 著者 | Roszak, A.W, Grinter, R, Cogdell, J.R, Walker, D. | | 登録日 | 2012-07-06 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | The Crystal Structure of the Lipid II-degrading Bacteriocin Syringacin M Suggests Unexpected Evolutionary Relationships between Colicin M-like Bacteriocins.
J.Biol.Chem., 287, 2012
|
|
2IVZ
 
 | | Structure of TolB in complex with a peptide of the colicin E9 T- domain | | 分子名称: | CALCIUM ION, COLICIN-E9, PROTEIN TOLB | | 著者 | Loftus, S.R, Walker, D, Mate, M.J, Bonsor, D.A, James, R, Moore, G.R, Kleanthous, C. | | 登録日 | 2006-06-23 | | 公開日 | 2006-08-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Competitive Recruitment of the Periplasmic Translocation Portal Tolb by a Natively Disordered Domain of Colicin E9
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
4LED
 
 | | The Crystal Structure of Pyocin L1 bound to D-rhamnose at 2.37 Angstroms | | 分子名称: | Pyocin L1, alpha-D-rhamnopyranose | | 著者 | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, C.J, Walker, D. | | 登録日 | 2013-06-25 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
4LEA
 
 | | The Crystal Structure of Pyocin L1 bound to D-mannose at 2.55 Angstroms | | 分子名称: | Pyocin L1, beta-D-mannopyranose | | 著者 | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, C.J, Walker, D. | | 登録日 | 2013-06-25 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
4LE7
 
 | | The Crystal Structure of Pyocin L1 at 2.09 Angstroms | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Pyocin L1 | | 著者 | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, R.J, Walker, D. | | 登録日 | 2013-06-25 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
4N58
 
 | | Crystal Structure of Pectocin M2 at 1.86 Angstroms | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Grinter, R, Roszak, A.W, Zeth, K, Cogdell, C.J, Walker, D. | | 登録日 | 2013-10-09 | | 公開日 | 2014-06-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structure of the atypical bacteriocin pectocin M2 implies a novel mechanism of protein uptake.
Mol.Microbiol., 93, 2014
|
|
4N59
 
 | | The Crystal Structure of Pectocin M2 at 2.3 Angstroms | | 分子名称: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Pectocin M2, ... | | 著者 | Zeth, K, Grinter, R, Roszak, A.W, Cogdell, R.J, Walker, D. | | 登録日 | 2013-10-09 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the atypical bacteriocin pectocin M2 implies a novel mechanism of protein uptake.
Mol.Microbiol., 93, 2014
|
|
4QKO
 
 | | The Crystal Structure of the Pyocin S2 Nuclease Domain, Immunity Protein Complex at 1.8 Angstroms | | 分子名称: | BROMIDE ION, MAGNESIUM ION, Pyocin-S2, ... | | 著者 | Grinter, R, Josts, I, Roszak, A.W, Cogdell, C.J, Walker, D. | | 登録日 | 2014-06-07 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Insights into pyocin S2
To be Published
|
|
6O2Z
 
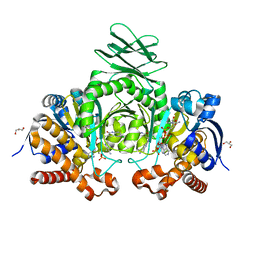 | | Crystal structure of IDH1 R132H mutant in complex with compound 32 | | 分子名称: | 6-{[(6-chloro-2-oxo-1,2-dihydroquinolin-3-yl)methyl]amino}-2-methylpyridine-3-carbonitrile, GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | 著者 | Toms, A.V, Lin, J. | | 登録日 | 2019-02-25 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery and Optimization of Quinolinone Derivatives as Potent, Selective, and Orally Bioavailable Mutant Isocitrate Dehydrogenase 1 (mIDH1) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6O2Y
 
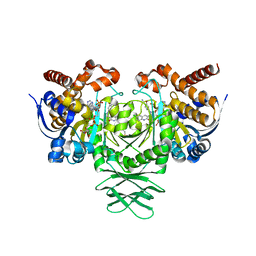 | | Crystal structure of IDH1 R132H mutant in complex with compound 24 | | 分子名称: | 4-{[(6-chloro-2-oxo-1,2-dihydroquinolin-3-yl)methyl]amino}-2-methoxybenzonitrile, BETA-MERCAPTOETHANOL, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | 著者 | Toms, A.V, Lin, J. | | 登録日 | 2019-02-25 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery and Optimization of Quinolinone Derivatives as Potent, Selective, and Orally Bioavailable Mutant Isocitrate Dehydrogenase 1 (mIDH1) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6OFS
 
 | |
6OFT
 
 | |
6OFR
 
 | |
6THK
 
 | |
