1Q4S
 
 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase complexed with CoA and 4-hydroxybenzoic acid | | Descriptor: | COENZYME A, P-HYDROXYBENZOIC ACID, Thioesterase | | Authors: | Thoden, J.B, Zhuang, Z, Dunaway-Mariano, D, Holden, H.M. | | Deposit date: | 2003-08-04 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. strain SU
J.Biol.Chem., 278, 2003
|
|
1Q4U
 
 | | Crystal structure of 4-hydroxybenzoyl CoA thioesterase from arthrobacter sp. strain SU complexed with 4-hydroxybenzyl CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYBENZYL COENZYME A, Thioesterase | | Authors: | Thoden, J.B, Zhuang, Z, Dunaway-Mariano, D, Holden, H.M. | | Deposit date: | 2003-08-04 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. strain SU
J.Biol.Chem., 278, 2003
|
|
1MMX
 
 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with D-fucose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, alpha-L-fucopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
5J63
 
 | | Crystal Structure of the N-terminal N-formyltransferase Domain (residues 1-306) of Escherichia coli Arna in Complex with UDP-Ara4N and Folinic Acid | | Descriptor: | (2R,3R,4S,5S)-5-amino-3,4-dihydroxytetrahydro-2H-pyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, (4aS,6S)-2-amino-6-{(E)-[(4-methylphenyl)imino]methyl}-4-oxo-4,6,7,8-tetrahydropteridine-5(4aH)-carbaldehyde, Bifunctional polymyxin resistance protein ArnA | | Authors: | Thoden, J.B, Genthe, N.A, Holden, H.M. | | Deposit date: | 2016-04-04 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Escherichia coli ArnA N-formyltransferase domain in complex with N(5) -formyltetrahydrofolate and UDP-Ara4N.
Protein Sci., 25, 2016
|
|
7US5
 
 | | X-ray crystal structure of GDP-D-glycero-D-manno-heptose 4,6-Dehydratase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-GLYCERO-D-MANNO-HEPTOSE 4,6-DEHYDRATASE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-04-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction Mechanism and Three-Dimensional Structure of GDP-d-glycero-alpha-d-manno-heptose 4,6-Dehydratase from Campylobacter jejuni.
Biochemistry, 61, 2022
|
|
4XD1
 
 | | X-ray structure of the N-formyltransferase QdtF from Providencia alcalifaciens, W305A mutant, in the presence of TDP-Qui3N and N5-THF | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Thoden, J.B, Woodford, C.R, Holden, H.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New Role for the Ankyrin Repeat Revealed by a Study of the N-Formyltransferase from Providencia alcalifaciens.
Biochemistry, 54, 2015
|
|
4XD0
 
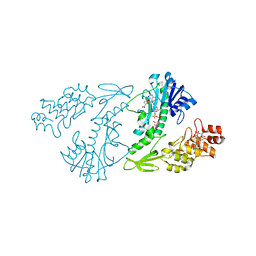 | | X-ray structure of the N-formyltransferase QdtF from Providencia alcalifaciens | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, CHLORIDE ION, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Thoden, J.B, Woodford, C.R, Holden, H.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New Role for the Ankyrin Repeat Revealed by a Study of the N-Formyltransferase from Providencia alcalifaciens.
Biochemistry, 54, 2015
|
|
4XCZ
 
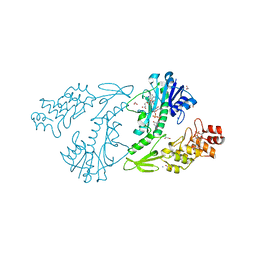 | | X-ray structure of the N-formyltransferase QdtF from Providencia alcalifaciens in complex with TDP-Qui3n and N5-THF | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Thoden, J.B, Woodford, C.R, Holden, H.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New Role for the Ankyrin Repeat Revealed by a Study of the N-Formyltransferase from Providencia alcalifaciens.
Biochemistry, 54, 2015
|
|
8DB5
 
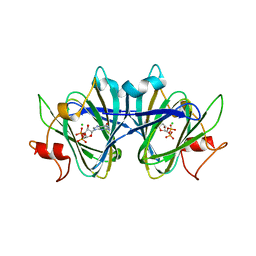 | | Crystal structure of the GDP-D-glycero-4-keto-d-lyxo-heptose-3,5-epimerase from Campylobacter jejuni, serotype HS:15 | | Descriptor: | CHLORIDE ION, GDP-D-glycero-4-keto-d-lyxo-heptose-3,5-epimerase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-14 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
8DAK
 
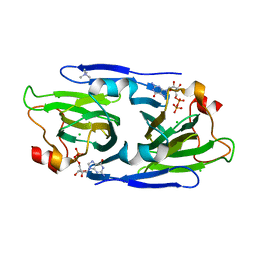 | | Crystal structure of the GDP-D-glycero-4-keto-d-lyxo-heptose-3-epimerase from Campylobacter jejuni, serotype HS:3 | | Descriptor: | CHLORIDE ION, GDP-D-glycero-4-keto-d-lyxo-heptose-3-epimerase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-13 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
8DCO
 
 | | Crystal structure of the GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase from Campylobacter jejuni, serotype HS:42 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase, ... | | Authors: | Thoden, J.B, Xiang, D.F, Ghosh, M.K, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
8DCL
 
 | | Crystal structure of the GDP-D-glycero-4-keto-D-lyxo-heptose-3-epimerase from campylobacter jejuni, serotype HS:23/36 | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-glycero-4-keto-D-lyxo-heptose-3-epimerase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-16 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
8V4H
 
 | | X-ray structure of the NADP-dependent reductase from Campylobacter jejuni responsible for the synthesis of CDP-glucitol in the presence of CDP-glucitol | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Putative nucleotide sugar dehydratase, ... | | Authors: | Thoden, J.B, Schumann, M.E, Holden, H.M, Raushel, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biosynthesis of Cytidine Diphosphate-6-d-Glucitol for the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 63, 2024
|
|
6VLO
 
 | | X-ray Structure of the R141 Sugar 4,6-dehydratase from Acanthamoeba polyphaga Minivirus | | Descriptor: | NICKEL (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative dTDP-D-glucose 4,6-dehydratase, ... | | Authors: | Thoden, J.B, Ferek, J.D, Holden, H.M. | | Deposit date: | 2020-01-24 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical analysis of a sugar 4,6-dehydratase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 29, 2020
|
|
4NV1
 
 | | Crystal structure of a 4-N formyltransferase from Francisella tularensis | | Descriptor: | Formyltransferase, PHOSPHATE ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Zimmer, A.L, Holden, H.M. | | Deposit date: | 2013-12-04 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of a sugar N-formyltransferase from Francisella tularensis.
Protein Sci., 23, 2014
|
|
5BSZ
 
 | | X-ray structure of the sugar N-methyltransferase KedS8 from Streptoalloteichus sp ATCC 53650 | | Descriptor: | 1,2-ETHANEDIOL, N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Thoden, J.B, Delvaux, N.A, Holden, H.M. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular architecture of KedS8, a sugar N-methyltransferase from Streptoalloteichus sp. ATCC 53650.
Protein Sci., 24, 2015
|
|
3Q2I
 
 | | Crystal structure of the WlbA dehydrognase from Chromobactrium violaceum in complex with NADH and UDP-GlcNAcA at 1.50 A resolution | | Descriptor: | (2S,3S,4R,5R,6R)-5-acetamido-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4-dihydroxy-oxane-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2010-12-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and Structural Characterization of WlbA from Bordetella pertussis and Chromobacterium violaceum: Enzymes Required for the Biosynthesis of 2,3-Diacetamido-2,3-dideoxy-d-mannuronic Acid.
Biochemistry, 50, 2011
|
|
3Q2K
 
 | | Crystal structure of the WlbA dehydrogenase from Bordetella pertussis in complex with NADH and UDP-GlcNAcA | | Descriptor: | (2S,3S,4R,5R,6R)-5-acetamido-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4-dihydroxy-oxane-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, oxidoreductase | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2010-12-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Biochemical and Structural Characterization of WlbA from Bordetella pertussis and Chromobacterium violaceum: Enzymes Required for the Biosynthesis of 2,3-Diacetamido-2,3-dideoxy-d-mannuronic Acid.
Biochemistry, 50, 2011
|
|
8SXW
 
 | | X-ray crystal structure of UDP- 2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27, D98N mutation, apo structure at pH 6 | | Descriptor: | CHLORIDE ION, SODIUM ION, UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase | | Authors: | Kroft, C.W, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
8SY0
 
 | | X-ray crystal structure of UDP- 2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27 in complex with its product UDP-2,3-diacetamido-2,3-dideoxy-d-mannuronic acid at pH 9 | | Descriptor: | (2~{S},3~{S},4~{R},5~{S},6~{R})-4,5-diacetamido-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxane-2-carboxylic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | McKnight, J.O, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
8SXY
 
 | | X-ray crystal structure of UDP- 2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27 in complex with its product UDP-2,3-diacetamido-2,3-dideoxy-d-mannuronic acid at pH 5 | | Descriptor: | (2~{S},3~{S},4~{R},5~{S},6~{R})-4,5-diacetamido-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxane-2-carboxylic acid, CHLORIDE ION, UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase | | Authors: | McKnight, J.O, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
8SYD
 
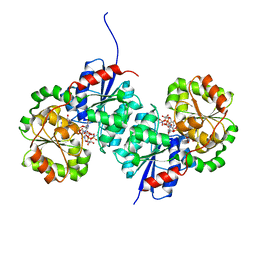 | | X-ray crystal structure of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27, D98N variant in the presence of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid and UDP-N-acetylglucosamine at pH 6 | | Descriptor: | (2~{S},3~{S},4~{R},5~{R},6~{R})-4,5-diacetamido-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxane-2-carboxylic acid, CHLORIDE ION, UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase, ... | | Authors: | Kroft, C.W, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-25 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
8SYE
 
 | | X-ray crystal structure of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27, D98N variant in the presence of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid and UDP at pH 6 | | Descriptor: | (2~{S},3~{S},4~{R},5~{R},6~{R})-4,5-diacetamido-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxane-2-carboxylic acid, CHLORIDE ION, UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase, ... | | Authors: | Jast, J.D.T, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-25 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
8SXV
 
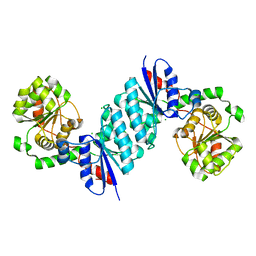 | | X-ray crystal structure of UDP- 2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27, apo form, pH 9 | | Descriptor: | CHLORIDE ION, UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase | | Authors: | McKnight, J.O, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
8SYH
 
 | | X-ray crystal structure of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27, D98N variant in the presence of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid and UDP at pH 8 | | Descriptor: | (2~{S},3~{S},4~{R},5~{R},6~{R})-4,5-diacetamido-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxane-2-carboxylic acid, CHLORIDE ION, UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase, ... | | Authors: | Jast, J.D.T, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-25 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
