8DWH
 
 | | CryoEM structure of Gq-coupled MRGPRX1 with ligand Compound-16 | | 分子名称: | Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Liu, Y, Cao, C, Fay, J.F, Roth, B.L. | | 登録日 | 2022-08-01 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Ligand recognition and allosteric modulation of the human MRGPRX1 receptor.
Nat.Chem.Biol., 19, 2023
|
|
8DWG
 
 | | CryoEM structure of Gq-coupled MRGPRX1 with peptide ligand BAM8-22 and positive allosteric modulator ML382 | | 分子名称: | 2-[(cyclopropanesulfonyl)amino]-N-(2-ethoxyphenyl)benzamide, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Liu, Y, Cao, C, Fay, J.F, Roth, B.L. | | 登録日 | 2022-08-01 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Ligand recognition and allosteric modulation of the human MRGPRX1 receptor.
Nat.Chem.Biol., 19, 2023
|
|
2BLS
 
 | |
4XGK
 
 | | Crystal structure of UDP-galactopyranose mutase from Corynebacterium diphtheriae in complex with 2-[4-(4-chlorophenyl)-7-(2-thienyl)-2-thia-5,6,8,9-tetrazabicyclo[4.3.0]nona-4,7,9-trien-3-yl]acetic | | 分子名称: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, UDP-galactopyranose mutase, ... | | 著者 | Wangkanont, K, Heroux, A, Forest, K.T, Kiessling, L.L. | | 登録日 | 2014-12-31 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.652 Å) | | 主引用文献 | Virtual Screening for UDP-Galactopyranose Mutase Ligands Identifies a New Class of Antimycobacterial Agents.
Acs Chem.Biol., 10, 2015
|
|
4XX9
 
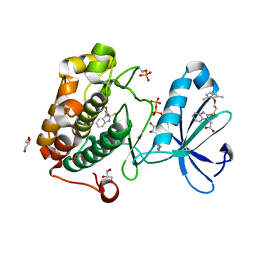 | |
7KQW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, methylated) | | 分子名称: | Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-17 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.93 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQP
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose (P43 crystal form) | | 分子名称: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-17 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (0.88 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQO
 
 | |
7KR0
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 100 K) | | 分子名称: | Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-18 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.77 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR1
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 310 K) | | 分子名称: | Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-18 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7M96
 
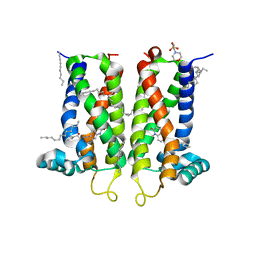 | | Bovine sigma-2 receptor bound to Z4857158944 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-({[(1S)-1-hydroxy-2-methyl-1-phenylpropan-2-yl]amino}methyl)-1-methyl-3,4-dihydroquinolin-2(1H)-one, ... | | 著者 | Alon, A, Kruse, A.C. | | 登録日 | 2021-03-30 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
7M95
 
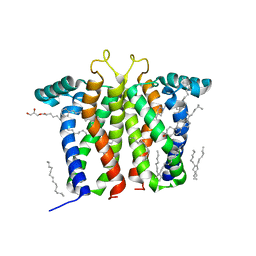 | | Bovine sigma-2 receptor bound to Z1241145220 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[1-(3-phenylpropyl)-1,2,3,6-tetrahydropyridin-4-yl]-1H-pyrrolo[2,3-b]pyridine, CHOLESTEROL, ... | | 著者 | Alon, A, Kruse, A.C. | | 登録日 | 2021-03-30 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
7MFI
 
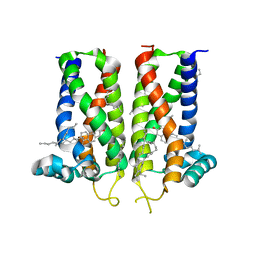 | | Bovine sigma-2 receptor bound to cholesterol | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHOLESTEROL, ... | | 著者 | Alon, A, Kruse, A.C. | | 登録日 | 2021-04-09 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
7M94
 
 | | Bovine sigma-2 receptor bound to Roluperidone | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Roluperidone, Sigma intracellular receptor 2 | | 著者 | Alon, A, Kruse, A.C. | | 登録日 | 2021-03-30 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
7M93
 
 | | Bovine sigma-2 receptor bound to PB28 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PB28, Sigma intracellular receptor 2 | | 著者 | Alon, A, Kruse, A.C. | | 登録日 | 2021-03-30 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
4JM8
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 2,6-diaminopyridine | | 分子名称: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Boyce, S.E, Fischer, M, Fish, I. | | 登録日 | 2013-03-13 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JMA
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 3-Fluorocatechol | | 分子名称: | 3-FLUOROBENZENE-1,2-DIOL, Cytochrome c peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Boyce, S.E, Fischer, M, Fish, I. | | 登録日 | 2013-03-13 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JM9
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 3-amino-1-methylpyridinium | | 分子名称: | 1-METHYL-1,6-DIHYDROPYRIDIN-3-AMINE, Cytochrome c peroxidase, IODIDE ION, ... | | 著者 | Boyce, S.E, Fischer, M, Fish, I. | | 登録日 | 2013-03-13 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JMW
 
 | |
4JM6
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 2,4-diaminopyrimidine | | 分子名称: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Boyce, S.E, Fischer, M, Fish, I. | | 登録日 | 2013-03-13 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4NVK
 
 | | Predicting protein conformational response in prospective ligand discovery. | | 分子名称: | Cytochrome c peroxidase, N~2~,N~2~-diethylquinazoline-2,4-diamine, PHOSPHATE ION, ... | | 著者 | Fischer, M, Fraser, J.S. | | 登録日 | 2013-12-05 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Incorporation of protein flexibility and conformational energy penalties in docking screens to improve ligand discovery.
Nat Chem, 6, 2014
|
|
4NVN
 
 | | Predicting protein conformational response in prospective ligand discovery | | 分子名称: | 2,3-dihydrobenzo[h][1,6]naphthyridin-4(1H)-one, Cytochrome c peroxidase, PHOSPHATE ION, ... | | 著者 | Fischer, M, Fraser, J.S. | | 登録日 | 2013-12-05 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Incorporation of protein flexibility and conformational energy penalties in docking screens to improve ligand discovery.
Nat Chem, 6, 2014
|
|
4NVB
 
 | |
4NVM
 
 | |
4NVE
 
 | |
