1RIS
 
 | | CRYSTAL STRUCTURE OF THE RIBOSOMAL PROTEIN S6 FROM THERMUS THERMOPHILUS | | Descriptor: | RIBOSOMAL PROTEIN S6 | | Authors: | Lindahl, M, Svensson, L.A, Liljas, A, Sedelnikova, S.E, Eliseikina, I.A, Fomenkova, N.P, Nevskaya, N, Nikonov, S.V, Garber, M.B, Muranova, T.A, Rykonova, A.I, Amons, R. | | Deposit date: | 1994-05-31 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the ribosomal protein S6 from Thermus thermophilus.
EMBO J., 13, 1994
|
|
5HML
 
 | | Crystal Structure of T5 D15 Protein Co-crystallized with Metal Ions | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Flemming, C.S, Feng, M, Sedelnikova, S.E, Zhang, J, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-16 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5HMM
 
 | | Crystal Structure of T5 D15 Protein Co-crystallized with Metal Ions | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exodeoxyribonuclease, ... | | Authors: | Flemming, C.S, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-16 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5HP4
 
 | | Crystal structure bacteriohage T5 D15 flap endonuclease (D155K) pseudo-enzyme-product complex with DNA and metal ions | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*AP*TP*CP*TP*AP*TP*AP*TP*GP*CP*CP*AP*TP*CP*GP*G)-3'), Exodeoxyribonuclease, ... | | Authors: | Almalki, F.A, Zhang, J, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
1SUL
 
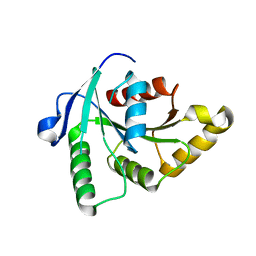 | | Crystal Structure of the apo-YsxC | | Descriptor: | GTP-binding protein YsxC | | Authors: | Ruzheinikov, S.N, Das, K.S, Sedelnikova, S.E, Baker, P.J, Artymiuk, P.J, Garcia-Lara, J, Foster, S.J, Rice, D.W. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the Open and Closed Conformations of the GTP-binding Protein YsxC from Bacillus subtilis.
J.Mol.Biol., 339, 2004
|
|
1SVI
 
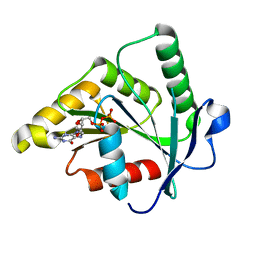 | | Crystal Structure of the GTP-binding protein YsxC complexed with GDP | | Descriptor: | GTP-binding protein YSXC, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Baker, P.J, Artymiuk, P.J, Garcia-Lara, J, Foster, S.J, Rice, D.W. | | Deposit date: | 2004-03-29 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of the Open and Closed Conformations of the GTP-binding Protein YsxC from Bacillus subtilis.
J.Mol.Biol., 339, 2004
|
|
1SVW
 
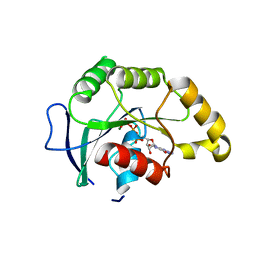 | | Crystal Structure of YsxC complexed with GMPPNP | | Descriptor: | GTP-binding protein YsxC, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Baker, P.J, Artymiuk, P.J, Garcia-Lara, J, Foster, S.J, Rice, D.W. | | Deposit date: | 2004-03-30 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the Open and Closed Conformations of the GTP-binding Protein YsxC from Bacillus subtilis.
J.Mol.Biol., 339, 2004
|
|
1X82
 
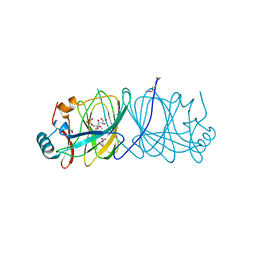 | | CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE FROM PYROCOCCUS FURIOSUS WITH BOUND 5-phospho-D-arabinonate | | Descriptor: | 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-17 | | Release date: | 2004-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
1X7N
 
 | | The crystal structure of Pyrococcus furiosus phosphoglucose isomerase with bound 5-phospho-D-arabinonate and Manganese | | Descriptor: | 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase, MANGANESE (II) ION | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-16 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
1X8E
 
 | | Crystal structure of Pyrococcus furiosus phosphoglucose isomerase free enzyme | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
1Z8R
 
 | | 2A cysteine proteinase from human coxsackievirus B4 (strain JVB / Benschoten / New York / 51) | | Descriptor: | Coxsackievirus B4 polyprotein, ZINC ION | | Authors: | Baxter, N.J, Roetzer, A, Liebig, H.D, Sedelnikova, S.E, Hounslow, A.M, Skern, T, Waltho, J.P. | | Deposit date: | 2005-03-31 | | Release date: | 2006-02-14 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of coxsackievirus B4 2A proteinase, an enyzme involved in the etiology of heart disease.
J.Virol., 80, 2006
|
|
1ZUJ
 
 | | The crystal structure of the Lactococcus lactis MG1363 DpsA protein | | Descriptor: | hypothetical protein Llacc01001955 | | Authors: | Stillman, T.J, Upadhyay, M, Norte, V.A, Sedelnikova, S.E, Carradus, M, Tzokov, S, Bullough, P.A, Shearman, C.A, Gasson, M.J, Williams, C.H, Artymiuk, P.J, Green, J. | | Deposit date: | 2005-05-31 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding.
Mol.Microbiol., 57, 2005
|
|
1ZS3
 
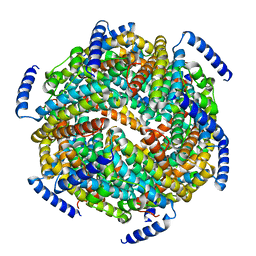 | | The crystal structure of the Lactococcus lactis MG1363 DpsB protein | | Descriptor: | Lactococcus lactis MG1363 DpsA | | Authors: | Stillman, T.J, Upadhyay, M, Norte, V.A, Sedelnikova, S.E, Carradus, M, Tzokov, S, Bullough, P.A, Shearman, C.A, Gasson, M.J, Williams, C.H, Artymiuk, P.J, Green, J. | | Deposit date: | 2005-05-23 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding.
Mol.Microbiol., 57, 2005
|
|
4MU4
 
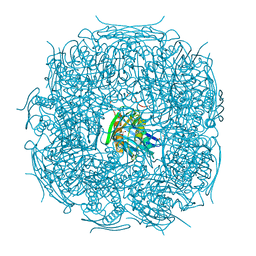 | | The form B structure of an E21Q catalytic mutant of A. thaliana IGPD2 in complex with Mn2+ and its substrate, 2R3S-IGP, to 1.41 A resolution | | Descriptor: | (2R,3S)-2,3-dihydroxy-3-(1H-imidazol-5-yl)propyl dihydrogen phosphate, 1,2-ETHANEDIOL, Imidazoleglycerol-phosphate dehydratase 2, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
4MU0
 
 | | The structure of wt A. thaliana IGPD2 in complex with Mn2+ and 1,2,4-triazole at 1.3 A resolution | | Descriptor: | 1,2,4-TRIAZOLE, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
1IGW
 
 | | Crystal Structure of the Isocitrate Lyase from the A219C mutant of Escherichia coli | | Descriptor: | Isocitrate lyase, MAGNESIUM ION, MERCURY (II) ION, ... | | Authors: | Britton, K.L, Abeysinghe, I.S.B, Baker, P.J, Barynin, V, Diehl, P, Langridge, S.J, McFadden, B.A, Sedelnikova, S.E, Stillman, T.J, Weeradechapon, K, Rice, D.W. | | Deposit date: | 2001-04-18 | | Release date: | 2001-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure and domain organization of Escherichia coli isocitrate lyase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1K30
 
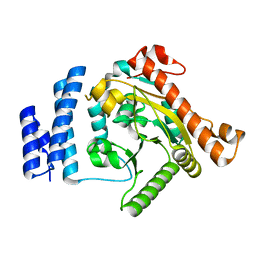 | | Crystal Structure Analysis of Squash (Cucurbita moschata) glycerol-3-phosphate (1)-acyltransferase | | Descriptor: | glycerol-3-phosphate acyltransferase | | Authors: | Turnbull, A.P, Rafferty, J.B, Sedelnikova, S.E, Slabas, A.R, Schierer, T.P, Kroon, J.T, Simon, J.W, Fawcett, T, Nishida, I, Murata, N, Rice, D.W. | | Deposit date: | 2001-10-01 | | Release date: | 2001-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of the structure, substrate specificity, and mechanism of squash glycerol-3-phosphate (1)-acyltransferase.
Structure, 9, 2001
|
|
1JQW
 
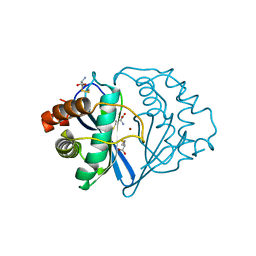 | | THE 2.3 ANGSTROM RESOLUTION STRUCTURE OF BACILLUS SUBTILIS LUXS/HOMOCYSTEINE COMPLEX | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Autoinducer-2 production protein luxS, ZINC ION | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-08-09 | | Release date: | 2001-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.2 A structure of a novel quorum-sensing protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
1JVI
 
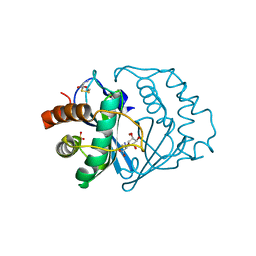 | | THE 2.2 ANGSTROM RESOLUTION STRUCTURE OF BACILLUS SUBTILIS LUXS/RIBOSILHOMOCYSTEINE COMPLEX | | Descriptor: | (2S)-2-amino-4-[[(2S,3S,4R,5R)-3,4,5-trihydroxyoxolan-2-yl]methylsulfanyl]butanoic acid, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Autoinducer-2 production protein luxS, ... | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-08-30 | | Release date: | 2001-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 1.2 A structure of a novel quorum-sensing protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
1Q8R
 
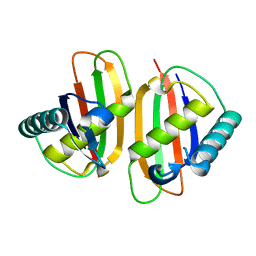 | | Structure of E.coli RusA Holliday junction resolvase | | Descriptor: | Crossover junction endodeoxyribonuclease rusA | | Authors: | Rafferty, J.B, Bolt, E.L, Muranova, T.A, Sedelnikova, S.E, Leonard, P, Pasquo, A, Baker, P.J, Rice, D.W, Sharples, G.J, Lloyd, R.G. | | Deposit date: | 2003-08-22 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | The structure of Escherichia coli RusA endonuclease reveals a new Holliday junction DNA binding fold
Structure, 11, 2003
|
|
1KTG
 
 | | Crystal Structure of a C. elegans Ap4A Hydrolase Binary Complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Diadenosine Tetraphosphate Hydrolase, HYDROXIDE ION, ... | | Authors: | Bailey, S, Sedelnikova, S.E, Blackburn, G.M, Abdelghany, H.M, Baker, P.J, McLennan, A.G, Rafferty, J.B. | | Deposit date: | 2002-01-16 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of diadenosine tetraphosphate hydrolase from Caenorhabditis elegans in free and binary complex forms
Structure, 10, 2002
|
|
1MIE
 
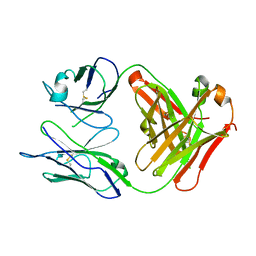 | | Crystal Structure Of The Fab Fragment of Esterolytic Antibody MS5-393 | | Descriptor: | IMMUNOGLOBULIN MS5-393 | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-23 | | Release date: | 2003-09-23 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1MJJ
 
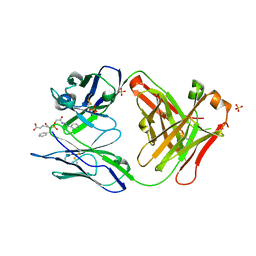 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX OF THE FAB FRAGMENT OF ESTEROLYTIC ANTIBODY MS6-12 AND A TRANSITION-STATE ANALOG | | Descriptor: | IMMUNOGLOBULIN MS6-12, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE, SULFATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-28 | | Release date: | 2003-09-23 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1MH5
 
 | | The Structure Of The Complex Of The Fab Fragment Of The Esterolytic Antibody MS6-164 and A Transition-State Analog | | Descriptor: | IMMUNOGLOBULIN MS6-164, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE, SULFATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-19 | | Release date: | 2003-09-23 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1MJ8
 
 | | High Resolution Crystal Structure Of The Fab Fragment of The Esterolytic Antibody MS6-126 | | Descriptor: | GLYCEROL, IMMUNOGLOBULIN MS6-126, PHOSPHATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-27 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
