8V8W
 
 | |
8V8Y
 
 | |
8V8X
 
 | |
9AZZ
 
 | |
8VW0
 
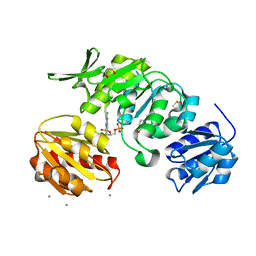 | |
8VW2
 
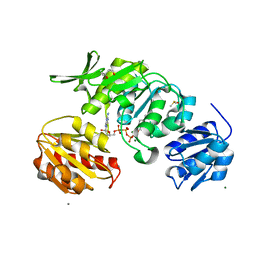 | |
8VW1
 
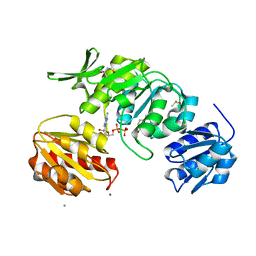 | |
4XK1
 
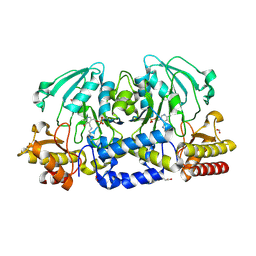 | |
4XWI
 
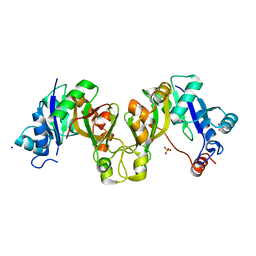 | |
6Q09
 
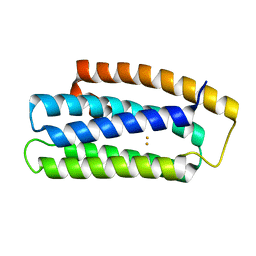 | |
5HL6
 
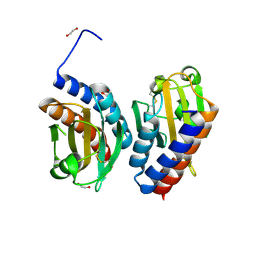 | |
6PZJ
 
 | |
5IF9
 
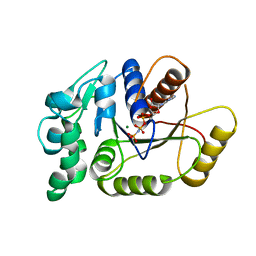 | |
8SAB
 
 | |
8SA8
 
 | |
8SAA
 
 | |
8SAD
 
 | |
8SBW
 
 | | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (Apo, orthorhombic form) | | Descriptor: | 2,3-dihydroxybenzoate-2,3-dehydrogenase, ACETATE ION, SULFATE ION | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2023-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (Apo, orthorhombic form)
To be published
|
|
8SBZ
 
 | |
8SA7
 
 | |
8SBY
 
 | | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (NAD and sulfate bound, hexagonal form) | | Descriptor: | 2,3-dihydroxybenzoate-2,3-dehydrogenase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2023-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (NAD and sulfate bound, hexagonal form)
To be published
|
|
8SBN
 
 | |
8SBV
 
 | |
8SC0
 
 | | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (NAD bound, orthorhombic form) | | Descriptor: | 2,3-dihydroxybenzoate-2,3-dehydrogenase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2023-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (NAD bound, orthorhombic form)
To be published
|
|
8SAE
 
 | |
