1H62
 
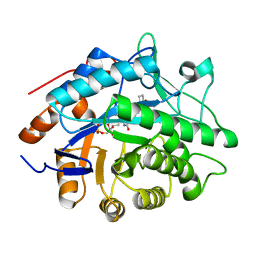 | | Structure of Pentaerythritol tetranitrate reductase in complex with 1,4-androstadien-3,17-dione | | Descriptor: | ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2001-06-04 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
1H60
 
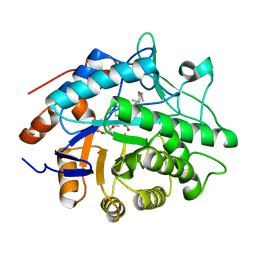 | |
2XXF
 
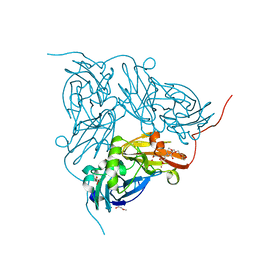 | | Cu metallated H254F mutant of nitrite reductase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hough, M.A, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2010-11-10 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
2YL3
 
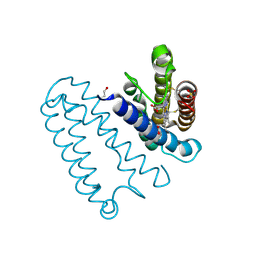 | | CYTOCHROME C PRIME FROM ALCALIGENES XYLOSOXIDANS: CARBON MONOOXIDE BOUND L16G VARIANT AT 1.04 A RESOLUTION - RESTRAINT REFINED | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C, ... | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-05-31 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2YLD
 
 | | RECOMBINANT NATIVE CYTOCHROME C PRIME FROM ALCALIGENES XYLOSOXIDANS: CARBON MONOOXIDE BOUND AT 1.25 A | | Descriptor: | ASCORBIC ACID, CARBON MONOXIDE, CYTOCHROME C', ... | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-06-02 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2YKZ
 
 | |
2YL0
 
 | |
2YL7
 
 | | CYTOCHROME C PRIME FROM ALCALIGENES XYLOSOXIDANS: AS ISOLATED L16G VARIANT AT 0.9 A RESOLUTION - RESTRAINT REFINEMENT | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-05-31 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2YLG
 
 | | CYTOCHROME C PRIME FROM ALCALIGENES XYLOSOXIDANS: ASCORBATE AND CARBON MONOOXIDE BOUND L16A VARIANT AT 1.05 A RESOLUTION | | Descriptor: | ASCORBIC ACID, CARBON MONOXIDE, CYTOCHROME C', ... | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-06-02 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2YLI
 
 | |
2YL1
 
 | |
6F53
 
 | |
6F4Y
 
 | |
6F54
 
 | |
6F50
 
 | |
6FOZ
 
 | |
6FOY
 
 | |
6FP1
 
 | | The crystal structure of P.fluorescens Kynurenine 3-monooxygenase (KMO) in complex with competitive inhibitor No. 1 | | Descriptor: | 2-(6-chloranyl-5,7-dimethyl-3-oxidanylidene-1,4-benzoxazin-4-yl)ethanoic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Levy, C.W, Leys, D. | | Deposit date: | 2018-02-08 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A brain-permeable inhibitor of the neurodegenerative disease target kynurenine 3-monooxygenase prevents accumulation of neurotoxic metabolites.
Commun Biol, 2, 2019
|
|
6FPH
 
 | | The crystal structure of P.fluorescens Kynurenine 3-monooxygenase (KMO) in complex with competitive inhibitor No. 1h | | Descriptor: | 6-chloranyl-5,7-dimethyl-4-(1~{H}-1,2,3,4-tetrazol-5-ylmethyl)-1,4-benzoxazin-3-one, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Levy, C.W, Leys, D. | | Deposit date: | 2018-02-09 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A brain-permeable inhibitor of the neurodegenerative disease target kynurenine 3-monooxygenase prevents accumulation of neurotoxic metabolites.
Commun Biol, 2, 2019
|
|
4ZA5
 
 | | Structure of A. niger Fdc1 with the prenylated-flavin cofactor in the iminium and ketimine forms. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 1-deoxy-5-O-phosphono-1-[(10aR)-2,2,3,4-tetramethyl-8,10-dioxo-1,2,8,9,10,10a-hexahydro-6H-indeno[1,7-ef]pyrimido[4,5-b][1,4]diazepin-6-yl]-D-ribitol, Fdc1, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4ZA4
 
 | | Structure of A. niger Fdc1 with the prenylated-flavin cofactor in the iminium form. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Fdc1, MANGANESE (II) ION, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4ZAF
 
 | |
4ZAY
 
 | | Structure of UbiX E49Q in complex with a covalent adduct between dimethylallyl monophosphate and reduced FMN | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
4ZAG
 
 | |
4ZAZ
 
 | | Structure of UbiX Y169F in complex with a covalent adduct formed between reduced FMN and dimethylallyl monophosphate | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, PHOSPHATE ION, SODIUM ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
