1KAH
 
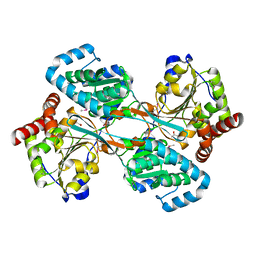 | | L-HISTIDINOL DEHYDROGENASE (HISD) STRUCTURE COMPLEXED WITH L-HISTIDINE (PRODUCT), ZN AND NAD (COFACTOR) | | 分子名称: | HISTIDINE, Histidinol dehydrogenase, ZINC ION | | 著者 | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J.D, Cygler, M. | | 登録日 | 2001-11-02 | | 公開日 | 2002-06-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KSL
 
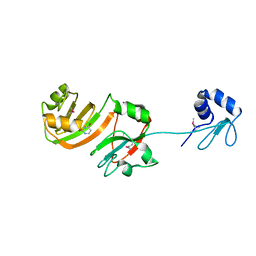 | | STRUCTURE OF RSUA | | 分子名称: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URACIL | | 著者 | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2002-01-13 | | 公開日 | 2002-04-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1KSV
 
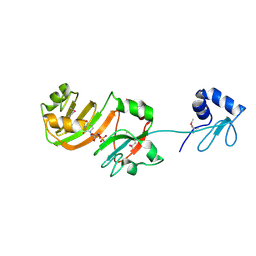 | | STRUCTURE OF RSUA | | 分子名称: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URIDINE-5'-MONOPHOSPHATE | | 著者 | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A. | | 登録日 | 2002-01-14 | | 公開日 | 2002-04-24 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1KSK
 
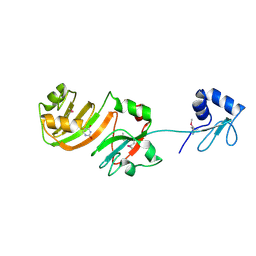 | | STRUCTURE OF RSUA | | 分子名称: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URACIL | | 著者 | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A. | | 登録日 | 2002-01-13 | | 公開日 | 2002-04-24 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
3LIP
 
 | |
6MXR
 
 | |
6MY4
 
 | | Crystal structure of the dimeric bH1-Fab variant [HC-Y33W,HC-D98M,HC-G99M,LC-S30bR] | | 分子名称: | 1,2-ETHANEDIOL, anti-VEGF-A Fab fragment bH1 heavy chain, anti-VEGF-A Fab fragment bH1 light chain | | 著者 | Shi, R, Picard, M.-E, Manenda, M. | | 登録日 | 2018-11-01 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Binding symmetry and surface flexibility mediate antibody self-association.
Mabs, 11, 2019
|
|
6MXS
 
 | | Crystal structure of the dimeric bH1-Fab variant [HC-Y33W,HC-D98F,HC-G99M] | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | 著者 | Shi, R, Picard, M.-E, Manenda, M.S. | | 登録日 | 2018-10-31 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Binding symmetry and surface flexibility mediate antibody self-association.
Mabs, 11, 2019
|
|
6MY5
 
 | |
1FC5
 
 | |
1LPS
 
 | | A STRUCTURAL BASIS FOR THE CHIRAL PREFERENCES OF LIPASES | | 分子名称: | (1S)-MENTHYL HEXYL PHOSPHONATE GROUP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Grochulski, P.G, Cygler, M.C. | | 登録日 | 1995-01-05 | | 公開日 | 1995-02-14 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | A Structural Basis for the Chiral Preferences of Lipases
J.Am.Chem.Soc., 116, 1994
|
|
1LPP
 
 | |
1LPO
 
 | |
1LPM
 
 | | A STRUCTURAL BASIS FOR THE CHIRAL PREFERENCES OF LIPASES | | 分子名称: | (1R)-MENTHYL HEXYL PHOSPHONATE GROUP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Grochulski, P.G, Cygler, M.C. | | 登録日 | 1995-01-06 | | 公開日 | 1995-04-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | A Structural Basis for the Chiral Preferences of Lipases
J.Am.Chem.Soc., 116, 1994
|
|
1LPN
 
 | |
1CRL
 
 | |
5LIP
 
 | | PSEUDOMONAS LIPASE COMPLEXED WITH RC-(RP, SP)-1,2-DIOCTYLCARBAMOYLGLYCERO-3-O-OCTYLPHOSPHONATE | | 分子名称: | CALCIUM ION, OCTYL-PHOSPHINIC ACID 1,2-BIS-OCTYLCARBAMOYLOXY-ETHYL ESTER, TRIACYL-GLYCEROL HYDROLASE | | 著者 | Lang, D.A, Dijkstra, B.W. | | 登録日 | 1997-09-02 | | 公開日 | 1998-08-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of the chiral selectivity of Pseudomonas cepacia lipase
Eur.J.Biochem., 254, 1998
|
|
2FLO
 
 | |
3HI4
 
 | |
1TRH
 
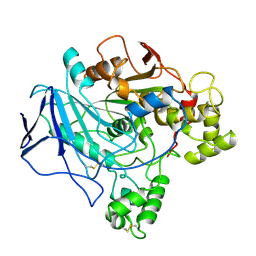 | |
1K75
 
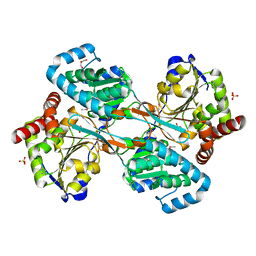 | | The L-histidinol dehydrogenase (hisD) structure implicates domain swapping and gene duplication. | | 分子名称: | GLYCEROL, L-histidinol dehydrogenase, SULFATE ION | | 著者 | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2001-10-18 | | 公開日 | 2002-02-27 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4LIP
 
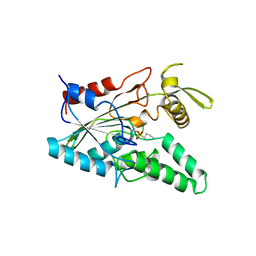 | |
1FG7
 
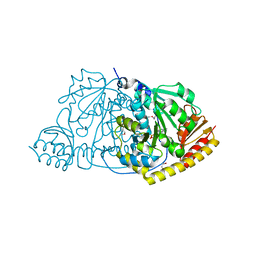 | |
1FG3
 
 | |
3EC3
 
 | |
