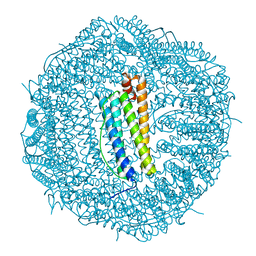
8A5T
 
 | |
2GIM
 
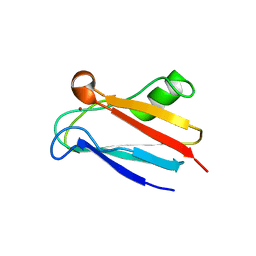 | |
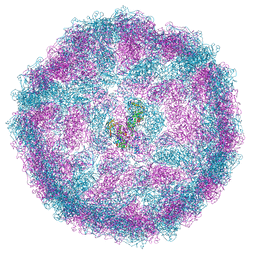
6SHT
 
 | | Molecular structure of mouse apoferritin resolved at 2.7 Angstroms with the Glacios cryo-microscope | | 分子名称: | FE (III) ION, Ferritin heavy chain, MAGNESIUM ION | | 著者 | Hamdi, F, Tueting, C, Semchonok, D, Kyrilis, F, Meister, A, Skalidis, I, Schmidt, L, Parthier, C, Stubbs, M.T, Kastritis, P.L. | | 登録日 | 2019-08-08 | | 公開日 | 2020-05-13 | | 最終更新日 | 2020-05-27 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | 2.7 angstrom cryo-EM structure of vitrified M. musculus H-chain apoferritin from a compact 200 keV cryo-microscope.
Plos One, 15, 2020
|
|
4QX1
 
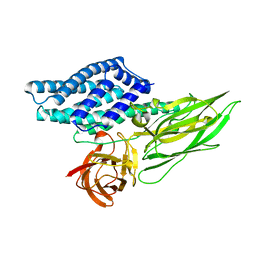 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the CrystFEL software suite | | 分子名称: | Pesticidal crystal protein cry3Aa | | 著者 | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | 登録日 | 2014-07-17 | | 公開日 | 2014-08-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QX3
 
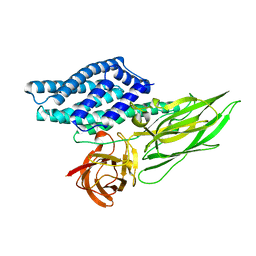 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the CrystFEL software suite | | 分子名称: | Pesticidal crystal protein cry3Aa | | 著者 | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | 登録日 | 2014-07-17 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QX0
 
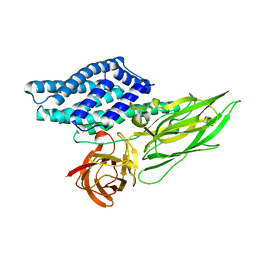 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the cctbx.xfel software suite | | 分子名称: | Pesticidal crystal protein cry3Aa | | 著者 | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | 登録日 | 2014-07-17 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QX2
 
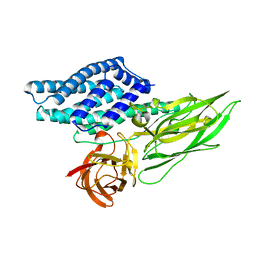 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the cctbx.xfel software suite | | 分子名称: | Pesticidal crystal protein cry3Aa | | 著者 | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | 登録日 | 2014-07-17 | | 公開日 | 2014-08-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8ADW
 
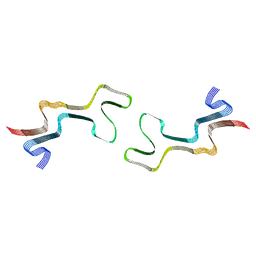 | | Lipidic alpha-synuclein fibril - polymorph L1C | | 分子名称: | Alpha-synuclein | | 著者 | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | 登録日 | 2022-07-11 | | 公開日 | 2022-10-12 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
8A4L
 
 | | Lipidic alpha-synuclein fibril - polymorph L2A | | 分子名称: | Alpha-synuclein | | 著者 | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | 登録日 | 2022-06-12 | | 公開日 | 2022-08-17 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
8ADU
 
 | | Lipidic alpha-synuclein fibril - polymorph L1A | | 分子名称: | Alpha-synuclein | | 著者 | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | 登録日 | 2022-07-11 | | 公開日 | 2022-10-12 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
8AEX
 
 | | Lipidic alpha-synuclein fibril - polymorph L3A | | 分子名称: | Alpha-synuclein | | 著者 | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | 登録日 | 2022-07-14 | | 公開日 | 2022-10-12 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
8ADS
 
 | | Lipidic alpha-synuclein fibril - polymorph L2B | | 分子名称: | Alpha-synuclein | | 著者 | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | 登録日 | 2022-07-11 | | 公開日 | 2022-10-12 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
8ADV
 
 | | Lipidic alpha-synuclein fibril - polymorph L1B | | 分子名称: | Alpha-synuclein | | 著者 | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | 登録日 | 2022-07-11 | | 公開日 | 2022-10-12 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
6YDY
 
 | |
4NP8
 
 | | Structure of an amyloid forming peptide VQIVYK from the second repeat region of tau (alternate polymorph) | | 分子名称: | Microtubule-associated protein tau | | 著者 | Landau, M, Eisenberg, D, Sawaya, M.R, Dannenberg, J, Kobko, N. | | 登録日 | 2013-11-20 | | 公開日 | 2013-12-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Molecular mechanisms for protein-encoded inheritance.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6BZM
 
 | | GFGNFGTS from low-complexity/FG repeat domain of Nup98, residues 116-123 | | 分子名称: | Nuclear pore complex protein Nup98-Nup96 | | 著者 | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Chong, L, Gonen, T, Eisenberg, D.S. | | 登録日 | 2017-12-24 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | 主引用文献 | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6BZP
 
 | | STGGYG from low-complexity domain of FUS, residues 77-82 | | 分子名称: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, RNA-binding protein FUS | | 著者 | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | 登録日 | 2017-12-25 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | 主引用文献 | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6BWZ
 
 | | SYSGYS from low-complexity domain of FUS, residues 37-42 | | 分子名称: | SYSGYS peptide from low-complexity domain of FUS | | 著者 | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Tamir, G, Eisenberg, D.S. | | 登録日 | 2017-12-15 | | 公開日 | 2018-04-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6BXV
 
 | | SYSSYGQS from low-complexity domain of FUS, residues 54-61 | | 分子名称: | FUS | | 著者 | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | 登録日 | 2017-12-19 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6BXX
 
 | | GYNGFG from low-complexity domain of hnRNPA1, residues 243-248 | | 分子名称: | hnRNPA1 | | 著者 | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | 登録日 | 2017-12-19 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
2Y3L
 
 | | Structure of segment MVGGVVIA from the amyloid-beta peptide (Ab, residues 35-42), alternate polymorph 2 | | 分子名称: | AMYLOID BETA A4 PROTEIN | | 著者 | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | 登録日 | 2010-12-21 | | 公開日 | 2011-11-02 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y3K
 
 | | Structure of segment MVGGVVIA from the amyloid-beta peptide (Ab, residues 35-42), alternate polymorph 1 | | 分子名称: | AMYLOID BETA A4 PROTEIN | | 著者 | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | 登録日 | 2010-12-21 | | 公開日 | 2011-11-02 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y3J
 
 | | Structure of segment AIIGLM from the amyloid-beta peptide (Ab, residues 30-35) | | 分子名称: | AMYLOID BETA A4 PROTEIN | | 著者 | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | 登録日 | 2010-12-21 | | 公開日 | 2011-11-02 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y29
 
 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph III | | 分子名称: | AMYLOID BETA A4 PROTEIN | | 著者 | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | 登録日 | 2010-12-14 | | 公開日 | 2011-10-26 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y2A
 
 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph I | | 分子名称: | ACETATE ION, AMYLOID BETA A4 PROTEIN | | 著者 | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | 登録日 | 2010-12-14 | | 公開日 | 2011-10-26 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
