1EDE
 
 | |
2DIJ
 
 | | COMPLEX OF A Y195F MUTANT CGTASE FROM B. CIRCULANS STRAIN 251 COMPLEXED WITH A MALTONONAOSE INHIBITOR AT PH 9.8 OBTAINED AFTER SOAKING THE CRYSTAL WITH ACARBOSE AND MALTOHEXAOSE | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Strokopytov, B.V, Knegtel, R.M.A, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 1998-05-27 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of cyclodextrin glycosyltransferase complexed with a maltononaose inhibitor at 2.6 angstrom resolution. Implications for product specificity.
Biochemistry, 35, 1996
|
|
2EDC
 
 | |
2EDA
 
 | |
1HDE
 
 | |
2DHE
 
 | |
2DHD
 
 | |
2DHC
 
 | |
2HVM
 
 | |
8BXL
 
 | | Patulin Synthase from Penicillium expansum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tjallinks, G, Boverio, A, Rozeboom, H.J, Fraaije, M.W. | | Deposit date: | 2022-12-09 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure elucidation and characterization of patulin synthase, insights into the formation of a fungal mycotoxin.
Febs J., 290, 2023
|
|
2CXG
 
 | | CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED TO THE INHIBITOR ACARBOSE | | Descriptor: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Strokopytov, B.V, Uitdehaag, J.C.M, Ruiterkamp, R, Dijkstra, B.W. | | Deposit date: | 1998-05-08 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of cyclodextrin glycosyltransferase complexed with acarbose. Implications for the catalytic mechanism of glycosidases.
Biochemistry, 34, 1995
|
|
2HAD
 
 | |
1DTU
 
 | | BACILLUS CIRCULANS STRAIN 251 CYCLODEXTRIN GLYCOSYLTRANSFERASE: A MUTANT Y89D/S146P COMPLEXED TO AN HEXASACCHARIDE INHIBITOR | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, PROTEIN (CYCLODEXTRIN GLYCOSYLTRANSFERASE), ... | | Authors: | Uitdehaag, J.C.M, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational design of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 to increase alpha-cyclodextrin production.
J.Mol.Biol., 296, 2000
|
|
1EO7
 
 | | BACILLUS CIRCULANS STRAIN 251 CYCLODEXTRIN GLYCOSYLTRANSFERASE IN COMPLEX WITH MALTOHEXAOSE | | Descriptor: | CALCIUM ION, PROTEIN (CYCLODEXTRIN GLYCOSYLTRANSFERASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 2000-03-22 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structures of maltohexaose and maltoheptaose bound at the donor sites of cyclodextrin glycosyltransferase give insight into the mechanisms of transglycosylation activity and cyclodextrin size specificity.
Biochemistry, 39, 2000
|
|
1EO5
 
 | |
1CXH
 
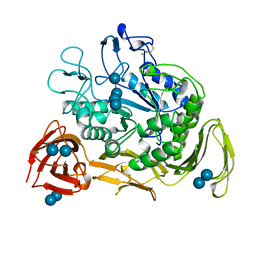 | | COMPLEX OF CGTASE WITH MALTOTETRAOSE AT ROOM TEMPERATURE AND PH 9.6 BASED ON DIFFRACTION DATA OF A CRYSTAL SOAKED WITH MALTOHEPTAOSE | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Knegtel, R.M.A, Strokopytov, B.V, Dijkstra, B.W. | | Deposit date: | 1995-07-31 | | Release date: | 1995-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystallographic studies of the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrates and products.
J.Biol.Chem., 270, 1995
|
|
1CXE
 
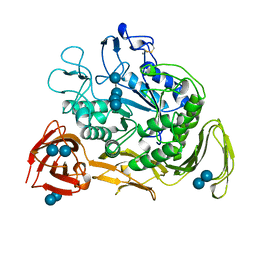 | | COMPLEX OF CGTASE WITH MALTOTETRAOSE AT ROOM TEMPERATURE AND PH 9.1 BASED ON DIFFRACTION DATA OF A CRYSTAL SOAKED WITH ALPHA-CYCLODEXTRIN | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Knegtel, R.M.A, Strokopytov, B.V, Dijkstra, B.W. | | Deposit date: | 1995-07-28 | | Release date: | 1995-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic studies of the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrates and products.
J.Biol.Chem., 270, 1995
|
|
1CIU
 
 | |
1CXI
 
 | |
1CXF
 
 | | COMPLEX OF A (D229N/E257Q) DOUBLE MUTANT CGTASE FROM BACILLUS CIRCULANS STRAIN 251 WITH MALTOTETRAOSE AT 120 K AND PH 9.1 OBTAINED AFTER SOAKING THE CRYSTAL WITH ALPHA-CYCLODEXTRIN | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Knegtel, R.M.A, Strokopytov, B.V, Dijkstra, B.W. | | Deposit date: | 1995-07-28 | | Release date: | 1995-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic studies of the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrates and products.
J.Biol.Chem., 270, 1995
|
|
1TCM
 
 | |
1GUV
 
 | | Structure of human chitotriosidase | | Descriptor: | 1,2-ETHANEDIOL, CHITOTRIOSIDASE | | Authors: | Von Moeller, H, Houston, D, Boot, R.G, Aerts, J.M.F.G, Van Aalten, D.M.F. | | Deposit date: | 2002-01-31 | | Release date: | 2003-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Human Chitotriosidase - Implications for Specific Inhibitor Design and Function of Mammalian Chitinase-Like Lectins
J.Biol.Chem., 277, 2002
|
|
1HVQ
 
 | | CRYSTAL STRUCTURES OF HEVAMINE, A PLANT DEFENCE PROTEIN WITH CHITINASE AND LYSOZYME ACTIVITY, AND ITS COMPLEX WITH AN INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEVAMINE A | | Authors: | Terwisscha Van Scheltinga, A.C, Kalk, K.H, Beintema, J.J, Dijkstra, B.W. | | Deposit date: | 1994-10-13 | | Release date: | 1995-12-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of hevamine, a plant defence protein with chitinase and lysozyme activity, and its complex with an inhibitor.
Structure, 2, 1994
|
|
1CGX
 
 | |
1CGW
 
 | |
