3UG7
 
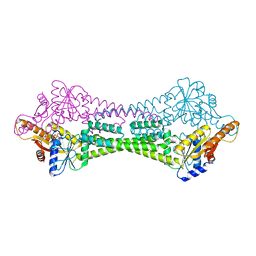 | | Crystal Structure of Get3 from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Suloway, C.J.M, Rome, M.E, Clemons Jr, W.M. | | Deposit date: | 2011-11-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Tail-anchor targeting by a Get3 tetramer: the structure of an archaeal homologue.
Embo J., 31, 2012
|
|
3UG6
 
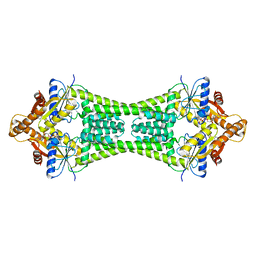 | | Crystal Structure of Get3 from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Suloway, C.J.M, Rome, M.E, Clemons Jr, W.M. | | Deposit date: | 2011-11-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tail-anchor targeting by a Get3 tetramer: the structure of an archaeal homologue.
Embo J., 31, 2012
|
|
4OAH
 
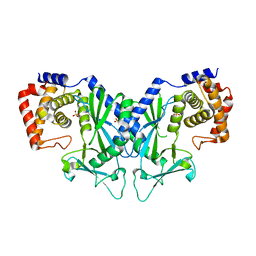 | |
5BWK
 
 | | 6.0 A Crystal structure of a Get3-Get4-Get5 intermediate complex from S.cerevisiae | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 4, Ubiquitin-like protein MDY2, ... | | Authors: | Gristick, H.B, Chartron, J.W, Clemons, W.M. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Mechanism of Assembly of a Substrate Transfer Complex during Tail-anchored Protein Targeting.
J.Biol.Chem., 290, 2015
|
|
5BW8
 
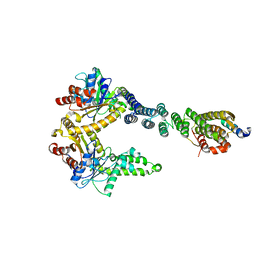 | | 2.8 A crystal structure of a Get3-Get4-Get5 intermediate complex from S.cerevisiae | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 4, Ubiquitin-like protein MDY2, ... | | Authors: | Gristick, H.B, Chartron, J.W, Clemons, W.M. | | Deposit date: | 2015-06-06 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of Assembly of a Substrate Transfer Complex during Tail-anchored Protein Targeting.
J.Biol.Chem., 290, 2015
|
|
4OAI
 
 | |
4OAF
 
 | |
4PWX
 
 | |
4OAG
 
 | |
