2ZBA
 
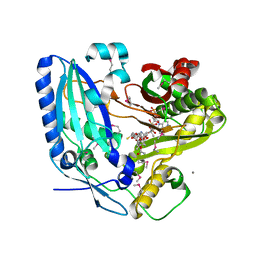 | | Crystal Structure of F. sporotrichioides TRI101 complexed with Coenzyme A and T-2 | | Descriptor: | 12,13-Epoxytrichothec-9-ene-3,4,8,15-tetrol-4,15-diacetate-8-isovalerate, CALCIUM ION, COENZYME A, ... | | Authors: | Garvey, G.S, McCormick, S.P, Rayment, I. | | Deposit date: | 2007-10-18 | | Release date: | 2007-12-11 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Characterization of the TRI101 Trichothecene 3-O-Acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum: KINETIC INSIGHTS TO COMBATING FUSARIUM HEAD BLIGHT
J.Biol.Chem., 283, 2008
|
|
3B30
 
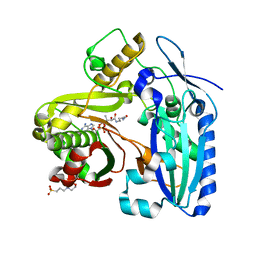 | | Crystal Structure of F. graminearum TRI101 complexed with Ethyl Coenzyme A | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Ethyl Coenzyme A, Trichothecene 3-O-acetyltransferase | | Authors: | Garvey, G.S, Rayment, I. | | Deposit date: | 2007-10-19 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Functional Characterization of the TRI101 Trichothecene 3-O-Acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum: KINETIC INSIGHTS TO COMBATING FUSARIUM HEAD BLIGHT
J.Biol.Chem., 283, 2008
|
|
3B2S
 
 | | Crystal Structure of F. graminearum TRI101 complexed with Coenzyme A and Deoxynivalenol | | Descriptor: | (3alpha,7alpha)-3,7,15-trihydroxy-12,13-epoxytrichothec-9-en-8-one, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, COENZYME A, ... | | Authors: | Garvey, G.S, Rayment, I. | | Deposit date: | 2007-10-19 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and Functional Characterization of the TRI101 Trichothecene 3-O-Acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum: KINETIC INSIGHTS TO COMBATING FUSARIUM HEAD BLIGHT
J.Biol.Chem., 283, 2008
|
|
3KAR
 
 | | THE MOTOR DOMAIN OF KINESIN-LIKE PROTEIN KAR3, A SACCHAROMYCES CEREVISIAE KINESIN-RELATED PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | Authors: | Gulick, A.M, Song, H, Endow, S, Rayment, I. | | Deposit date: | 1997-11-26 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of the yeast Kar3 motor domain complexed with Mg.ADP to 2.3 A resolution.
Biochemistry, 37, 1998
|
|
1HXP
 
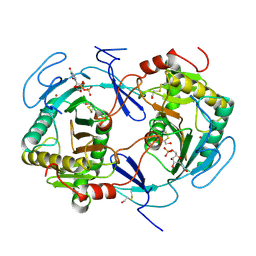 | | NUCLEOTIDE TRANSFERASE | | Descriptor: | BETA-MERCAPTOETHANOL, FE (III) ION, HEXOSE-1-PHOSPHATE URIDYLYLTRANSFERASE, ... | | Authors: | Wedekind, J.E, Frey, P.A, Rayment, I. | | Deposit date: | 1995-06-09 | | Release date: | 1996-11-08 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of galactose-1-phosphate uridylyltransferase from Escherichia coli at 1.8 A resolution.
Biochemistry, 34, 1995
|
|
3MYH
 
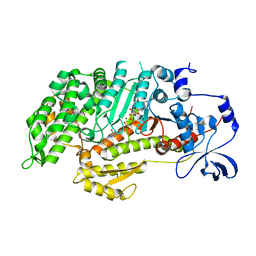 | | Insights into the Importance of Hydrogen Bonding in the Gamma-Phosphate Binding Pocket of Myosin: Structural and Functional Studies of Ser236 | | Descriptor: | (-)-1-PHENYL-1,2,3,4-TETRAHYDRO-4-HYDROXYPYRROLO[2,3-B]-7-METHYLQUINOLIN-4-ONE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Frye, J.J, Klenchin, V.A, Bagshaw, C.R, Rayment, I. | | Deposit date: | 2010-05-10 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Insights into the importance of hydrogen bonding in the gamma-phosphate binding pocket of myosin: structural and functional studies of serine 236
Biochemistry, 49, 2010
|
|
3MYL
 
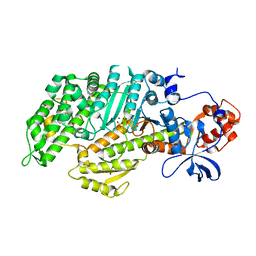 | | Insights into the Importance of Hydrogen Bonding in the Gamma-Phosphate Binding Pocket of Myosin: Structural and Functional Studies of Ser236 | | Descriptor: | MAGNESIUM ION, Myosin-2 heavy chain, PYROPHOSPHATE 2- | | Authors: | Frye, J.J, Klenchin, V.A, Bagshaw, C.R, Rayment, I. | | Deposit date: | 2010-05-10 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the importance of hydrogen bonding in the gamma-phosphate binding pocket of myosin: structural and functional studies of serine 236
Biochemistry, 49, 2010
|
|
3MTU
 
 | | Structure of the Tropomyosin Overlap Complex from Chicken Smooth Muscle | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Capsid assembly scaffolding protein,Tropomyosin alpha-1 chain, ... | | Authors: | Klenchin, V.A, Frye, J, Rayment, I. | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diversity of N-terminal recognition .
Biochemistry, 49, 2010
|
|
3MUD
 
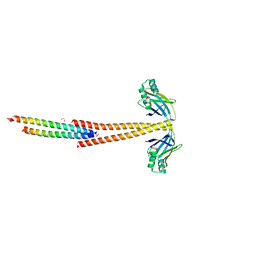 | | Structure of the Tropomyosin Overlap Complex from Chicken Smooth Muscle | | Descriptor: | 1,2-ETHANEDIOL, DNA repair protein XRCC4,Tropomyosin alpha-1 chain, SULFATE ION, ... | | Authors: | Klenchin, V.A, Frye, J, Rayment, I. | | Deposit date: | 2010-05-02 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diversity of N-terminal recognition .
Biochemistry, 49, 2010
|
|
3MYK
 
 | | Insights into the Importance of Hydrogen Bonding in the Gamma-Phosphate Binding Pocket of Myosin: Structural and Functional Studies of Ser236 | | Descriptor: | (-)-1-PHENYL-1,2,3,4-TETRAHYDRO-4-HYDROXYPYRROLO[2,3-B]-7-METHYLQUINOLIN-4-ONE, MAGNESIUM ION, Myosin-2 heavy chain, ... | | Authors: | Frye, J.J, Klenchin, V.A, Bagshaw, C.R, Rayment, I. | | Deposit date: | 2010-05-10 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Insights into the importance of hydrogen bonding in the gamma-phosphate binding pocket of myosin: structural and functional studies of serine 236
Biochemistry, 49, 2010
|
|
1JCT
 
 | | Glucarate Dehydratase, N341L mutant Orthorhombic Form | | Descriptor: | D-GLUCARATE, Glucarate Dehydratase, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-06-11 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in the active site of D-glucarate dehydratase from Escherichia coli.
Biochemistry, 40, 2001
|
|
3OA7
 
 | | Structure of the C-terminal domain of Cnm67, a core component of the spindle pole body of Saccharomyces cerevisiae | | Descriptor: | Head morphogenesis protein, Chaotic nuclear migration protein 67 fusion protein | | Authors: | Klenchin, V.A, Frye, J.J, Rayment, I. | | Deposit date: | 2010-08-04 | | Release date: | 2011-03-23 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function analysis of the C-terminal domain of CNM67, a core component of the Saccharomyces cerevisiae spindle pole body.
J.Biol.Chem., 286, 2011
|
|
5TMB
 
 | | Crystal structure of Os79 from O. sativa in complex with UDP. | | Descriptor: | Glycosyltransferase, Os79, URIDINE-5'-DIPHOSPHATE | | Authors: | Wetterhorn, K.M, Newmister, S.A, Caniza, R.K, Busman, M, McCormick, S.P, Berthiller, F, Adam, G, Rayment, I. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of Os79 (Os04g0206600) from Oryza sativa: A UDP-glucosyltransferase Involved in the Detoxification of Deoxynivalenol.
Biochemistry, 55, 2016
|
|
5TME
 
 | | Crystal structure of Os79 from O. sativa in complex with UDP. | | Descriptor: | Glycosyltransferase, Os79, URIDINE-5'-DIPHOSPHATE | | Authors: | Wetterhorn, K.M, Newmister, S.A, Caniza, R.K, Busman, M, McCormick, S.P, Berthiller, F, Adam, G, Rayment, I. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-02 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of Os79 (Os04g0206600) from Oryza sativa: A UDP-glucosyltransferase Involved in the Detoxification of Deoxynivalenol.
Biochemistry, 55, 2016
|
|
5TMD
 
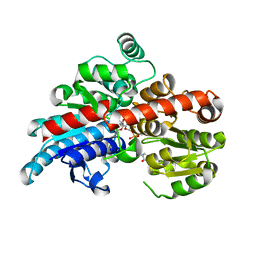 | | Crystal structure of Os79 from O. sativa in complex with U2F and trichothecene. | | Descriptor: | Glycosyltransferase, Os79, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE, ... | | Authors: | Wetterhorn, K, Newmister, S.A, Caniza, R.K, Busman, M, McCormick, S.P, Berthiller, F, Adam, G, Rayment, I. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Os79 (Os04g0206600) from Oryza sativa: A UDP-glucosyltransferase Involved in the Detoxification of Deoxynivalenol.
Biochemistry, 55, 2016
|
|
5WLZ
 
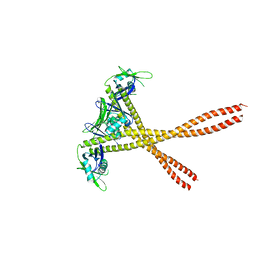 | | Crystal Structure of Amino Acids 1677-1758 of Human Beta Cardiac Myosin Fused to Xrcc4 | | Descriptor: | DNA repair protein XRCC4,Myosin-7 | | Authors: | Andreas, M.P, Ajay, G, Gellings, J, Rayment, I. | | Deposit date: | 2017-07-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design considerations in coiled-coil fusion constructs for the structural determination of a problematic region of the human cardiac myosin rod.
J. Struct. Biol., 200, 2017
|
|
5WLQ
 
 | | Crystal Structure of Amino Acids 1677-1755 of Human Beta Cardiac Myosin Fused to Gp7 and Eb1 | | Descriptor: | Capsid assembly scaffolding protein,Myosin-7,Microtubule-associated protein RP/EB family member 1, SULFATE ION, trimethylamine oxide | | Authors: | Andreas, M.P, Ajay, G, Gellings, J, Rayment, I. | | Deposit date: | 2017-07-27 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Design considerations in coiled-coil fusion constructs for the structural determination of a problematic region of the human cardiac myosin rod.
J. Struct. Biol., 200, 2017
|
|
5WME
 
 | | Crystal Structure of Amino Acids 1729-1786 of Human Beta Cardiac Myosin Fused to Gp7 as Anti-Parallel Four-Helix Bundle | | Descriptor: | Capsid assembly scaffolding protein,Myosin-7 | | Authors: | Andreas, M.P, Ajay, G, Gellings, J, Rayment, I. | | Deposit date: | 2017-07-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design considerations in coiled-coil fusion constructs for the structural determination of a problematic region of the human cardiac myosin rod.
J. Struct. Biol., 200, 2017
|
|
5JV3
 
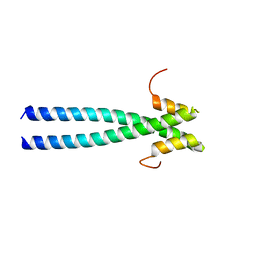 | | The neck-linker and alpha 7 helix of Homo sapiens Eg5 fused to EB1 | | Descriptor: | CALCIUM ION, Chimera protein of Kinesin-like protein KIF11 and Microtubule-associated protein RP/EB family member 1 | | Authors: | Phillips, R.K, Rayment, I. | | Deposit date: | 2016-05-10 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Family-specific Kinesin Structures Reveal Neck-linker Length Based on Initiation of the Coiled-coil.
J.Biol.Chem., 291, 2016
|
|
5JVS
 
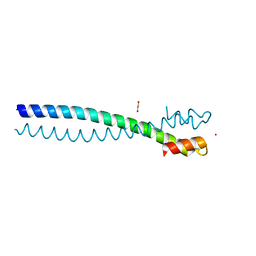 | |
5JVU
 
 | |
5JVR
 
 | |
5JVM
 
 | | The neck-linker and alpha 7 helix of Mus musculus KIF3C | | Descriptor: | Chimera protein of Kinesin-like protein KIF3C and Microtubule-associated protein RP/EB family member 1, MAGNESIUM ION | | Authors: | Phillips, R.K, Rayment, I. | | Deposit date: | 2016-05-11 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.567 Å) | | Cite: | Family-specific Kinesin Structures Reveal Neck-linker Length Based on Initiation of the Coiled-coil.
J.Biol.Chem., 291, 2016
|
|
5JVP
 
 | | The neck-linker and alpha 7 helix of Homo sapiens CENP-E | | Descriptor: | CADMIUM ION, Chimera protein of Centromere-associated protein E and Microtubule-associated protein RP/EB family member 1 | | Authors: | Phillips, R.K, Rayment, I. | | Deposit date: | 2016-05-11 | | Release date: | 2016-08-03 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Family-specific Kinesin Structures Reveal Neck-linker Length Based on Initiation of the Coiled-coil.
J.Biol.Chem., 291, 2016
|
|
5JX1
 
 | |
