4UW9
 
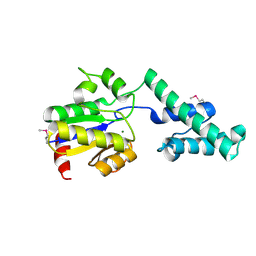 | |
4CMR
 
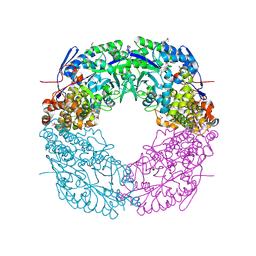 | | The crystal structure of novel exo-type maltose-forming amylase(Py04_0872) from Pyrococcus sp. ST04 | | 分子名称: | GLYCOSYL HYDROLASE/DEACETYLASE FAMILY PROTEIN | | 著者 | Park, K.-H, Jung, J.-H, Park, C.-S, Woo, E.-J. | | 登録日 | 2014-01-17 | | 公開日 | 2014-10-22 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Features Underlying the Selective Cleavage of a Novel Exo-Type Maltose-Forming Amylase from Pyrococcus Sp. St04
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2WSK
 
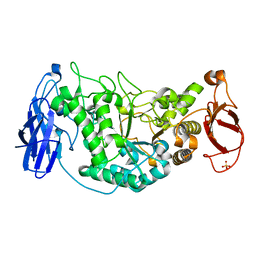 | | Crystal structure of Glycogen Debranching Enzyme GlgX from Escherichia coli K-12 | | 分子名称: | GLYCOGEN DEBRANCHING ENZYME, SULFATE ION | | 著者 | Song, H.-N, Park, J.-T, Jung, T.-Y, Park, K.-H, Woo, E.-J. | | 登録日 | 2009-09-08 | | 公開日 | 2010-09-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural Rationale for the Short Branched Substrate Specificity of the Glycogen Debranching Enzyme Glgx.
Proteins, 78, 2010
|
|
2WC7
 
 | | Crystal structure of Nostoc Punctiforme Debranching Enzyme(NPDE)(Acarbose soaked) | | 分子名称: | ALPHA AMYLASE, CATALYTIC REGION | | 著者 | Dumbrepatil, A.-B, Song, H.-N, Choi, J.-H, Park, K.-H, Woo, E.-J. | | 登録日 | 2009-03-10 | | 公開日 | 2009-09-29 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structural Features of the Nostoc Punctiforme Debranching Enzyme Reveal the Basis of its Mechanism and Substrate Specificity.
Proteins, 78, 2010
|
|
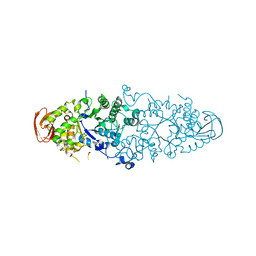
4AEF
 
 | | THE CRYSTAL STRUCTURE OF THERMOSTABLE AMYLASE FROM THE PYROCOCCUS | | 分子名称: | NEOPULLULANASE (ALPHA-AMYLASE II) | | 著者 | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Yang, S.-J, Park, K.-H, Woo, E.-J. | | 登録日 | 2012-01-10 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | A Novel Domain Arrangement in a Monomeric Cyclodextrin-Hydrolyzing Enzyme from the Hyperthermophile Pyrococcus Furiosus.
Biochim.Biophys.Acta, 1834, 2013
|
|
1PQR
 
 | | Solution Conformation of alphaA-Conotoxin EIVA | | 分子名称: | Alpha-A-conotoxin EIVA | | 著者 | Chi, S.-W, Park, K.-H, Suk, J.-E, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | 登録日 | 2003-06-18 | | 公開日 | 2003-11-04 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Conformation of alphaA-conotoxin EIVA, a Potent Neuromuscular Nicotinic Acetylcholine Receptor Antagonist from Conus ermineus
J.Biol.Chem., 278, 2003
|
|
4BBW
 
 | | The crystal structure of Sialidase VPI 5482 (BTSA) from Bacteroides thetaiotaomicron | | 分子名称: | SIALIDASE (NEURAMINIDASE) | | 著者 | Park, K.-H, Song, H.-N, Jung, T.-Y, Lee, M.-H, Woo, E.-J. | | 登録日 | 2012-09-28 | | 公開日 | 2013-08-14 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Biochemical Characterization of the Broad Substrate Specificity of Bacteroides Thetaiotaomicron Commensal Sialidase.
Biochim.Biophys.Acta, 1834, 2013
|
|
2VNC
 
 | | Crystal structure of Glycogen Debranching enzyme TreX from Sulfolobus solfataricus | | 分子名称: | GLYCOGEN OPERON PROTEIN GLGX | | 著者 | Song, H.-N, Yoon, S.-M, Cha, H, Park, K.-T, Woo, E.-J. | | 登録日 | 2008-02-04 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
2VR5
 
 | | Crystal structure of Trex from Sulfolobus Solfataricus in complex with acarbose intermediate and glucose | | 分子名称: | 4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranosyl)-beta-D-glucopyranose, GLYCEROL, GLYCOGEN OPERON PROTEIN GLGX, ... | | 著者 | Song, H.-N, Yoon, S.-M, Lee, S.-J, Cha, H.-J, Park, K.-H, Woo, E.-J. | | 登録日 | 2008-03-26 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
2VUY
 
 | | Crystal structure of Glycogen Debranching exzyme TreX from Sulfolobus solfatarius | | 分子名称: | GLYCOGEN OPERON PROTEIN GLGX | | 著者 | Song, H.-N, Yoon, S.-M, Cha, H.-J, Park, K.-H, Woo, E.-J. | | 登録日 | 2008-06-02 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
1DG2
 
 | | SOLUTION CONFORMATION OF A-CONOTOXIN AUIB | | 分子名称: | A-CONOTOXIN AUIB | | 著者 | Cho, J.-H, Mok, K.H, Olivera, B.M, McIntosh, J.M, Park, K.-H, Han, K.-H. | | 登録日 | 1999-11-23 | | 公開日 | 2000-02-25 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nuclear magnetic resonance solution conformation of alpha-conotoxin AuIB, an alpha(3)beta(4) subtype-selective neuronal nicotinic acetylcholine receptor antagonist.
J.Biol.Chem., 275, 2000
|
|
1EA9
 
 | | Cyclomaltodextrinase | | 分子名称: | CYCLOMALTODEXTRINASE | | 著者 | Cho, H.-S, Kim, M.-S, Oh, B.-H. | | 登録日 | 2001-07-12 | | 公開日 | 2002-06-06 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Cyclomaltodextrinase, Neopullulanase, and Maltogenic Amylase are Nearly Indistinguishable from Each Other
J.Biol.Chem., 277, 2002
|
|
1GVI
 
 | | Thermus maltogenic amylase in complex with beta-CD | | 分子名称: | Cycloheptakis-(1-4)-(alpha-D-glucopyranose), MALTOGENIC AMYLASE | | 著者 | Kim, M.-S, Kim, J.-I, Oh, B.-H. | | 登録日 | 2002-02-14 | | 公開日 | 2002-06-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Cyclomaltodextrinase, Neopullulanase, and Maltogenic Amylase are Nearly Indistinguishable from Each Other
J.Biol.Chem., 277, 2002
|
|
5FRU
 
 | |
5FRW
 
 | |
5FRY
 
 | |
5FRV
 
 | |
5FRZ
 
 | |
5FRX
 
 | |
