3LLQ
 
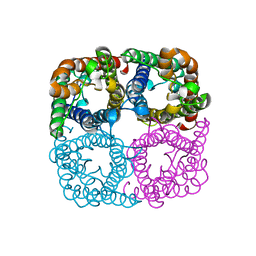 | |
3K3G
 
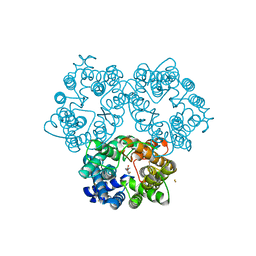 | | Crystal Structure of the Urea Transporter from Desulfovibrio Vulgaris Bound to 1,3-dimethylurea | | Descriptor: | 1,3-dimethylurea, GOLD ION, Urea transporter | | Authors: | Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a bacterial homologue of the kidney urea transporter.
Nature, 462, 2009
|
|
3TX3
 
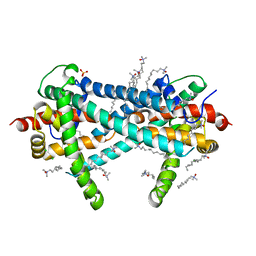 | | CysZ, a putative sulfate permease | | Descriptor: | CHLORIDE ION, LAURYL DIMETHYLAMINE-N-OXIDE, SULFATE ION, ... | | Authors: | Assur, Z, Liu, Q, Hendrickson, W.A, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2011-09-22 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CysZ, a putative sulfate permease
To be Published
|
|
3K3F
 
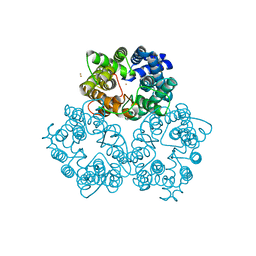 | |
3M78
 
 | |
3M7L
 
 | |
3M74
 
 | |
3M7E
 
 | |
3M77
 
 | |
3M7C
 
 | |
3M71
 
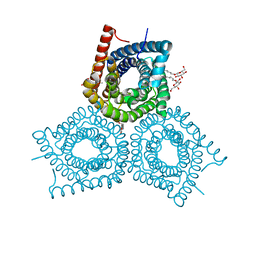 | | Crystal Structure of Plant SLAC1 homolog TehA | | Descriptor: | Tellurite resistance protein tehA homolog, octyl beta-D-glucopyranoside | | Authors: | Chen, Y.-H, Hu, L, Punta, M, Bruni, R, Hillerich, B, Kloss, B, Rost, B, Love, J, Siegelbaum, S.A, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2010-03-16 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Homologue structure of the SLAC1 anion channel for closing stomata in leaves.
Nature, 467, 2010
|
|
3ME1
 
 | |
3M75
 
 | |
3M76
 
 | |
3M73
 
 | |
3M7B
 
 | |
3KLY
 
 | |
3KLZ
 
 | |
3M6E
 
 | |
3M72
 
 | |
3M70
 
 | |
