7CG3
 
 | | Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum) | | 分子名称: | Heat shock protein 104 | | 著者 | Inoue, Y, Hanazono, Y, Noi, K, Kawamoto, A, Kimatsuka, M, Harada, R, Takeda, K, Iwamasa, N, Shibata, K, Noguchi, K, Shigeta, Y, Namba, K, Ogura, T, Miki, K, Shinohara, K, Yohda, M. | | 登録日 | 2020-06-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 29, 2021
|
|
5GV8
 
 | |
5GV7
 
 | |
6JGH
 
 | | Crystal structure of the F99S/M153T/V163A/T203I variant of GFP at 0.94 A | | 分子名称: | CHLORIDE ION, Green fluorescent protein | | 著者 | Eki, H, Tai, Y, Takaba, K, Hanazono, Y, Miki, K, Takeda, K. | | 登録日 | 2019-02-14 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.94 Å) | | 主引用文献 | Subatomic resolution X-ray structures of green fluorescent protein.
Iucrj, 6, 2019
|
|
6JGJ
 
 | | Crystal structure of the F99S/M153T/V163A/E222Q variant of GFP at 0.78 A | | 分子名称: | Green fluorescent protein, MAGNESIUM ION | | 著者 | Takaba, K, Tai, Y, Hanazono, Y, Miki, K, Takeda, K. | | 登録日 | 2019-02-14 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.78 Å) | | 主引用文献 | Subatomic resolution X-ray structures of green fluorescent protein.
Iucrj, 6, 2019
|
|
6JGI
 
 | | Crystal structure of the S65T/F99S/M153T/V163A variant of GFP at 0.85 A | | 分子名称: | Green fluorescent protein | | 著者 | Tai, Y, Takaba, K, Hanazono, Y, Miki, K, Takeda, K. | | 登録日 | 2019-02-14 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Subatomic resolution X-ray structures of green fluorescent protein.
Iucrj, 6, 2019
|
|
1WMF
 
 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (oxidized form, 1.73 angstrom) | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, GLYCEROL, ... | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | 登録日 | 2004-07-08 | | 公開日 | 2004-09-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
1WME
 
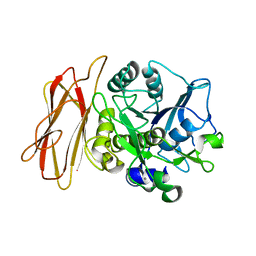 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (1.50 angstrom, 293 K) | | 分子名称: | CALCIUM ION, protease | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | 登録日 | 2004-07-08 | | 公開日 | 2004-09-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
1WMD
 
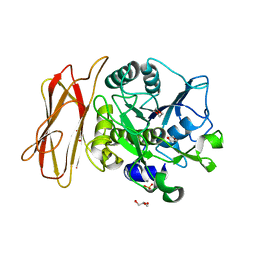 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (1.30 angstrom, 100 K) | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, GLYCEROL, ... | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | 登録日 | 2004-07-08 | | 公開日 | 2004-09-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
6KL1
 
 | | Crystal structure of the S65T/F99S/M153T/V163A variant of non-deuterated GFP at pD 8.5 | | 分子名称: | Green fluorescent protein | | 著者 | Tai, Y, Takaba, K, Hanazono, Y, Dao, H.A, Miki, K, Takeda, K. | | 登録日 | 2019-07-28 | | 公開日 | 2019-12-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.851 Å) | | 主引用文献 | X-ray crystallographic studies on the hydrogen isotope effects of green fluorescent protein at sub-angstrom resolutions
Acta Crystallogr.,Sect.D, 75, 2019
|
|
7E6P
 
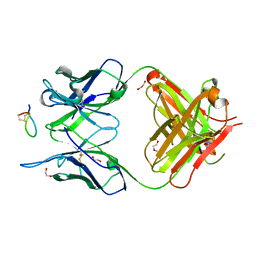 | | Fab-amyloid beta fragment complex | | 分子名称: | 1,2-ETHANEDIOL, Amyloid beta fragment with an intramolecular disulfide bond at positions 17 and 28, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kita, A, Irie, K, Irie, Y, Miki, K. | | 登録日 | 2021-02-23 | | 公開日 | 2022-01-05 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Characterization of a Conformation-Restricted Amyloid beta Peptide and Immunoreactivity of Its Antibody in Human AD brain.
Acs Chem Neurosci, 12, 2021
|
|
7BXV
 
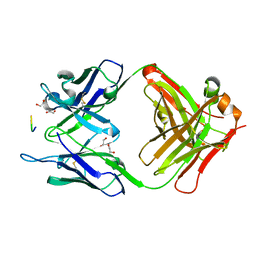 | | 11A1 antibody-peptide complex | | 分子名称: | 1,2-ETHANEDIOL, Fab of the 11A1 antibody H chain, Fab of the 11A1 antibody L chain, ... | | 著者 | Irie, K, Irie, Y, Kita, A, Miki, K. | | 登録日 | 2020-04-21 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | APOE epsilon 4 allele advances the age-dependent decline of amyloid beta clearance in the human cortex.
Biorxiv, 2022
|
|
5HWA
 
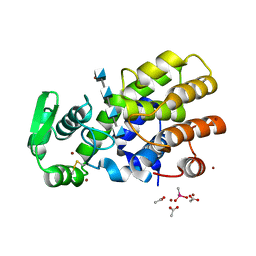 | | Crystal Structure of MH-K1 chitosanase in substrate-bound form | | 分子名称: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CACODYLATE ION, ... | | 著者 | Suzuki, M, Saito, A, Ando, A, Miki, K, Saito, J. | | 登録日 | 2016-01-29 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Crystal structure of the GH-46 subclass III chitosanase from Bacillus circulans MH-K1 in complex with chitotetraose
Biomed.Biochim.Acta, 1868, 2024
|
|
1JFL
 
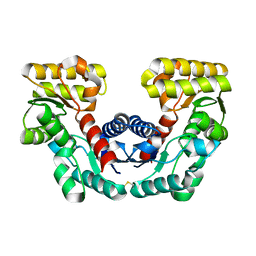 | | CRYSTAL STRUCTURE DETERMINATION OF ASPARTATE RACEMASE FROM AN ARCHAEA | | 分子名称: | ASPARTATE RACEMASE | | 著者 | Liu, L.J, Iwata, K, Kita, A, Kawarabayasi, Y, Yohda, M, Miki, K. | | 登録日 | 2001-06-21 | | 公開日 | 2002-06-05 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of aspartate racemase from Pyrococcus horikoshii OT3 and its implications for molecular mechanism of PLP-independent racemization.
J.Mol.Biol., 319, 2002
|
|
1WE0
 
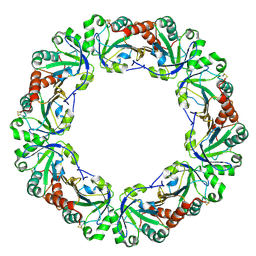 | | Crystal structure of peroxiredoxin (AhpC) from Amphibacillus xylanus | | 分子名称: | AMMONIUM ION, alkyl hydroperoxide reductase C | | 著者 | Kitano, K, Kita, A, Hakoshima, T, Niimura, Y, Miki, K. | | 登録日 | 2004-05-21 | | 公開日 | 2005-03-29 | | 最終更新日 | 2018-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of decameric peroxiredoxin (AhpC) from Amphibacillus xylanus
Proteins, 59, 2005
|
|
6IED
 
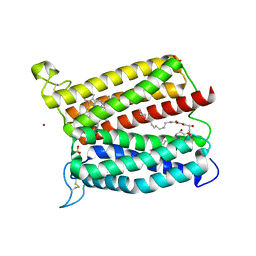 | | Crystal structure of heme A synthase from Bacillus subtilis | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Heme A synthase, ... | | 著者 | Niwa, S, Takeda, K, Kosugi, M, Tsutsumi, E, Miki, K. | | 登録日 | 2018-09-13 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of heme A synthase fromBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1HNL
 
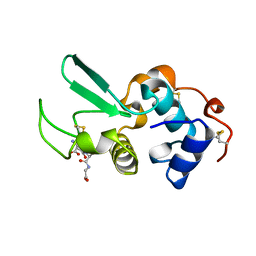 | |
7Y3J
 
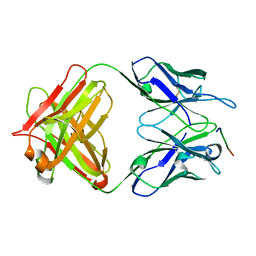 | | 24B3 antibody-peptide complex | | 分子名称: | 24B3 Heavy chain, 24B3 Light chain, ALA-LEU-VAL-PHE-PHE-ALA-PRO-ALA-VAL-GLY-SER | | 著者 | Irie, K, Irie, Y, Kita, A, Miki, K. | | 登録日 | 2022-06-11 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis of the 24B3 antibody against the toxic conformer of amyloid beta with a turn at positions 22 and 23.
Biochem.Biophys.Res.Commun., 621, 2022
|
|
4GA6
 
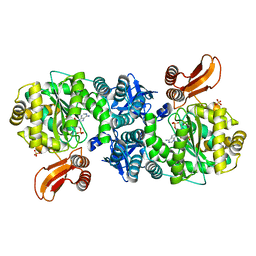 | | Crystal structure of AMP phosphorylase C-terminal deletion mutant in complex with substrates | | 分子名称: | ADENOSINE MONOPHOSPHATE, Putative thymidine phosphorylase, SULFATE ION | | 著者 | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | 登録日 | 2012-07-25 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
4GA4
 
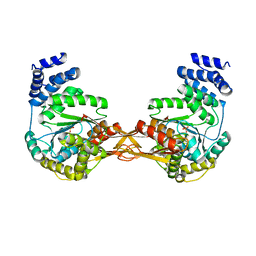 | | Crystal structure of AMP phosphorylase N-terminal deletion mutant | | 分子名称: | PHOSPHATE ION, Putative thymidine phosphorylase | | 著者 | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | 登録日 | 2012-07-25 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.51 Å) | | 主引用文献 | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
4G9I
 
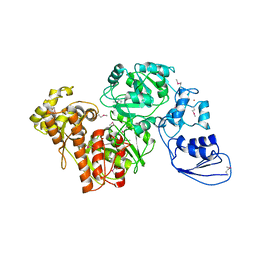 | | Crystal structure of T.kodakarensis HypF | | 分子名称: | Hydrogenase maturation protein HypF, ZINC ION | | 著者 | Tominaga, T, Watanabe, S, Matsumi, R, Atomi, H, Imanaka, T, Miki, K. | | 登録日 | 2012-07-24 | | 公開日 | 2012-10-24 | | 実験手法 | X-RAY DIFFRACTION (4.5 Å) | | 主引用文献 | Structure of the [NiFe]-hydrogenase maturation protein HypF from Thermococcus kodakarensis KOD1.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4GA5
 
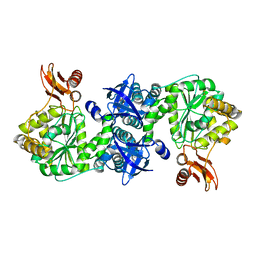 | | Crystal structure of AMP phosphorylase C-terminal deletion mutant in the apo-form | | 分子名称: | Putative thymidine phosphorylase | | 著者 | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | 登録日 | 2012-07-25 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
5HEE
 
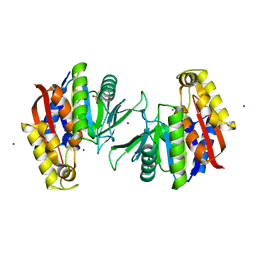 | | Crystal structure of the TK2203 protein | | 分子名称: | GLYCEROL, Putative uncharacterized protein, TK2203 protein, ... | | 著者 | Nishitani, Y, Miki, K. | | 登録日 | 2016-01-06 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Crystal structure of the TK2203 protein from Thermococcus kodakarensis, a putative extradiol dioxygenase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5HWS
 
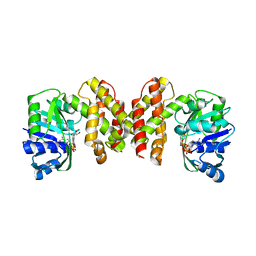 | |
5IJA
 
 | |
