7GRJ
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-6 | | Descriptor: | (5-chloro-1-benzothiophen-3-yl)methanol, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS3
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-26 | | Descriptor: | (6-phenylpyridin-3-yl)methanamine, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRU
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-17 | | Descriptor: | 3-(4-chlorophenyl)-1-methyl-1H-pyrazol-5-amine, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRR
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-14 | | Descriptor: | 3C-like proteinase nsp5, 5-(3-cyclohexylprop-1-yn-1-yl)pyridine-3-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRH
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-4 | | Descriptor: | 3C-like proteinase nsp5, 5-chloropyridin-3-ol, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRV
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-18 | | Descriptor: | (2S)-2-(2-fluorophenyl)-1,3-thiazolidin-4-one, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS0
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-23 | | Descriptor: | (pyridin-2-yl)(quinolin-2-yl)methanone, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS5
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-28 | | Descriptor: | (3R)-5-fluoro-3-hydroxy-1,3-dihydro-2H-indol-2-one, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRS
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-15 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRG
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-3 | | Descriptor: | 3,5-dichloropyridin-4-amine, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRQ
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-13 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRY
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-21 | | Descriptor: | 1-(3,5-dichlorophenyl)pyrrolidine-2,5-dione, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRF
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-2 | | Descriptor: | 3C-like proteinase nsp5, 5-bromopyridin-3-amine, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRZ
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-22 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N,N-dimethyl-2-[(naphthalen-2-yl)oxy]acetamide, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRE
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-1 | | Descriptor: | 3C-like proteinase nsp5, 4-[3-(trifluoromethyl)-1H-pyrazol-5-yl]pyridine, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A.M. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
5RDY
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 55 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R14
 
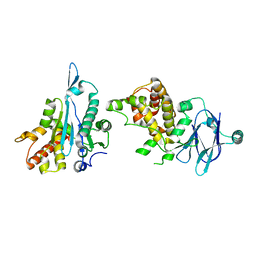 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 19, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R1I
 
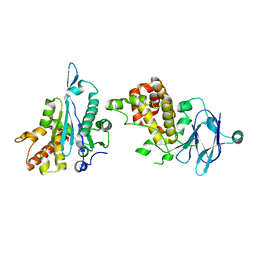 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 33, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R20
 
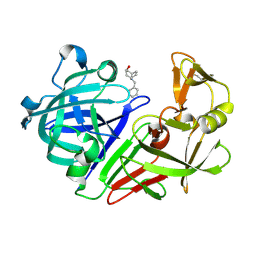 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry E06, DMSO-free | | Descriptor: | (3-endo)-8-benzyl-8-azabicyclo[3.2.1]octan-3-ol, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.009 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2G
 
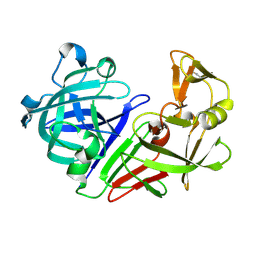 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 03, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (0.998 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2X
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 21, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.129 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3E
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 38, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.008 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3V
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 55, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.118 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RBT
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library C11b | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RCA
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library G08b | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
