5RU7
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000003591110 | | Descriptor: | 2,5-DIMETHYL-PYRIMIDIN-4-YLAMINE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUM
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000008861082 | | Descriptor: | 3-(3-oxo-3,4-dihydroquinoxalin-2-yl)propanoic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RV3
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000057162 | | Descriptor: | (5-methoxy-1H-indol-3-yl)acetic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVM
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000157088 | | Descriptor: | 4-HYDROXYBENZAMIDE, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2I60
 
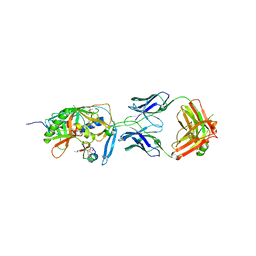 | | Crystal structure of [Phe23]M47, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 GP120 envelope glycoprotein and anti-HIV-1 antibody 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B HEAVY CHAIN, ANTIBODY 17B LIGHT CHAIN, ... | | Authors: | Huang, C.-C, Kwong, P.D. | | Deposit date: | 2006-08-26 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Combinatorial optimization of a CD4-mimetic miniprotein and cocrystal structures with HIV-1 gp120 envelope glycoprotein.
J.Mol.Biol., 382, 2008
|
|
2J3F
 
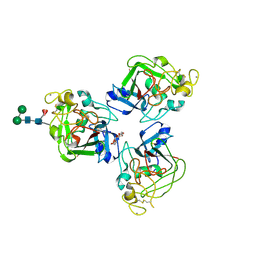 | | L-ficolin complexed to N-acetyl-D-galactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-08-21 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2J60
 
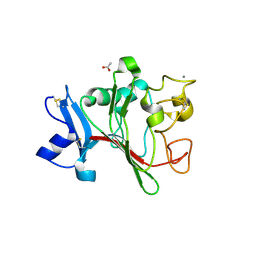 | | H-ficolin complexed to D-fucose | | Descriptor: | ACETATE ION, CALCIUM ION, FICOLIN-3, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-09-21 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2J0H
 
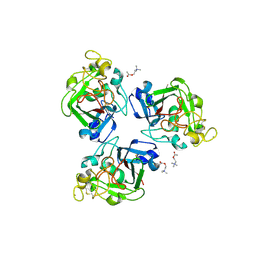 | | L-ficolin complexed to acetyl-choline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE, CALCIUM ION, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-08-03 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2J0Y
 
 | | L-ficolin complexed to b-1,3-D-glucan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-08-08 | | Release date: | 2007-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2J3O
 
 | | L-ficolin complexed to N-acetyl-D-glucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-08-22 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2J5Z
 
 | | H-ficolin complexed to galactose | | Descriptor: | ACETATE ION, CALCIUM ION, FICOLIN-3, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-09-21 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2J0G
 
 | | L-ficolin complexed to N-acetyl-mannosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-mannopyranose, CALCIUM ION, FICOLIN-2, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-08-03 | | Release date: | 2007-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2J1G
 
 | | L-ficolin complexed to N-acetyl-cystein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-08-11 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2J2P
 
 | | L-ficolin complexed to N-acetyl-cystein (150mM) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FICOLIN-2, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-08-17 | | Release date: | 2007-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2J61
 
 | |
2J3G
 
 | | L-ficolin | | Descriptor: | CALCIUM ION, FICOLIN-2, O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-08-21 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2J3U
 
 | | L-ficolin complexed to galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-08-23 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2J64
 
 | | H-ficolin | | Descriptor: | CALCIUM ION, FICOLIN-3 | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-09-25 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2C5W
 
 | | PENICILLIN-BINDING PROTEIN 1A (PBP-1A) ACYL-ENZYME COMPLEX (CEFOTAXIME) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | 1,2-ETHANEDIOL, CEFOTAXIME, C3' cleaved, ... | | Authors: | Contreras-Martel, C, Job, V, Di Guilmi, A.-M, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2005-11-02 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 1A (Pbp1A) Reveals a Mutational Hotspot Implicated in Beta-Lactam Resistance in Streptococcus Pneumoniae.
J.Mol.Biol., 355, 2006
|
|
2C6W
 
 | | PENICILLIN-BINDING PROTEIN 1A (PBP-1A) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1A, ZINC ION | | Authors: | Contreras-Martel, C, Job, V, Di Guilmi, A.-M, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2005-11-14 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 1A (Pbp1A) Reveals a Mutational Hotspot Implicated in Beta-Lactam Resistance in Streptococcus Pneumoniae.
J.Mol.Biol., 355, 2006
|
|
5W4L
 
 | | Crystal structure of the non-neutralizing and ADCC-potent C11-like antibody N12-i3 in complex with HIV-1 clade A/E gp120, the CD4 mimetic M48U1, and the antibody N5-i5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody N12-i3 Fab heavy chain, Antibody N12-i3 light chain, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Targeting the Late Stage of HIV-1 Entry for Antibody-Dependent Cellular Cytotoxicity: Structural Basis for Env Epitopes in the C11 Region.
Structure, 25, 2017
|
|
