6PTL
 
 | |
2LFN
 
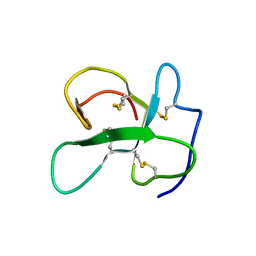 | | Identification of the key regions that drive functional amyloid formation by the fungal hydrophobin EAS | | 分子名称: | Hydrophobin | | 著者 | Macindoe, I, Kwan, A.H, Morris, V.K, Mackay, J.P, Sunde, M. | | 登録日 | 2011-07-06 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Self-assembly of functional, amphipathic amyloid monolayers by the fungal hydrophobin EAS
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6BI6
 
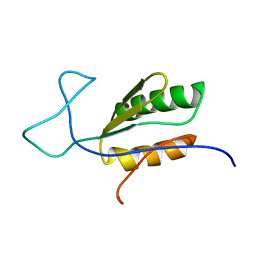 | | Solution NMR structure of uncharacterized protein YejG | | 分子名称: | Uncharacterized protein YejG | | 著者 | Mohanty, B, Finn, T.J, Macindoe, I, Zhong, J, Patrick, W.M, Mackay, J.P. | | 登録日 | 2017-11-01 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The uncharacterized bacterial protein YejG has the same architecture as domain III of elongation factor G.
Proteins, 87, 2019
|
|
2K6A
 
 | |
4BWH
 
 | |
