2R62
 
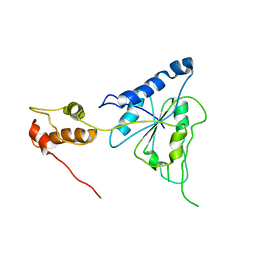 | | Crystal structure of Helicobacter pylori ATP dependent protease, FtsH | | 分子名称: | Cell division protease ftsH homolog | | 著者 | Kim, S.H, Kang, G.B, Song, H.-E, Park, S.J, Bae, M.-H, Eom, S.H. | | 登録日 | 2007-09-05 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural studies on Helicobacter pyloriATP-dependent protease, FtsH
J.SYNCHROTRON RADIAT., 15, 2008
|
|
2R65
 
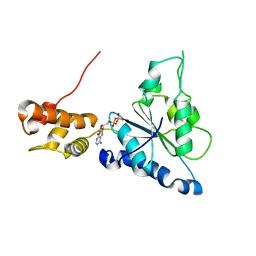 | | Crystal structure of Helicobacter pylori ATP dependent protease, FtsH ADP complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division protease ftsH homolog | | 著者 | Kim, S.H, Kang, G.B, Song, H.-E, Park, S.J, Bae, M.-H, Eom, S.H. | | 登録日 | 2007-09-05 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural studies on Helicobacter pyloriATP-dependent protease, FtsH
J.SYNCHROTRON RADIAT., 15, 2008
|
|
7VTG
 
 | | Pseudouridine bound structure of Pseudouridine kinase (PUKI) S30A mutant from Escherichia coli strain B | | 分子名称: | 5-[(2~{S},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1~{H}-pyrimidine-2,4-dione, Pseudouridine kinase | | 著者 | Kim, S.H, Rhee, S. | | 登録日 | 2021-10-29 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.89859128 Å) | | 主引用文献 | Substrate-binding loop interactions with pseudouridine trigger conformational changes that promote catalytic efficiency of pseudouridine kinase PUKI.
J.Biol.Chem., 298, 2022
|
|
7VVA
 
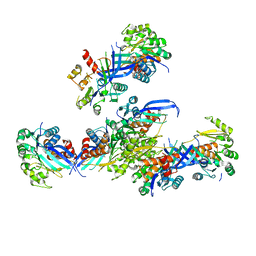 | | Pseudouridine bound structure of Pseudouridine kinase (PUKI) from Escherichia coli strain B | | 分子名称: | 5-[(2~{S},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1~{H}-pyrimidine-2,4-dione, Pseudouridine kinase | | 著者 | Kim, S.H, Rhee, S. | | 登録日 | 2021-11-05 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.75029182 Å) | | 主引用文献 | Substrate-binding loop interactions with pseudouridine trigger conformational changes that promote catalytic efficiency of pseudouridine kinase PUKI.
J.Biol.Chem., 298, 2022
|
|
7VTD
 
 | |
7VTF
 
 | |
7VTE
 
 | |
2ECR
 
 | | Crystal structure of the ligand-free form of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | 分子名称: | flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | 著者 | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | 登録日 | 2007-02-13 | | 公開日 | 2008-01-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
7DP1
 
 | |
7DP0
 
 | |
7DP2
 
 | |
2ED4
 
 | | Crystal structure of flavin reductase HpaC complexed with FAD and NAD | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygenae | | 著者 | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | 登録日 | 2007-02-14 | | 公開日 | 2008-01-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
2ECU
 
 | | Crystal structure of flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | 分子名称: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, DODECAETHYLENE GLYCOL, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygnease | | 著者 | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | 登録日 | 2007-02-14 | | 公開日 | 2008-01-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
7C1Z
 
 | |
7C1X
 
 | |
7C1Y
 
 | | Pseudouridine and ADP bound structure of Pseudouridine kinase (PUKI) from Arabidopsis thaliana | | 分子名称: | 5-[(2~{S},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1~{H}-pyrimidine-2,4-dione, ADENOSINE-5'-DIPHOSPHATE, PfkB-like carbohydrate kinase family protein, ... | | 著者 | Kim, S.H, Rhee, S. | | 登録日 | 2020-05-06 | | 公開日 | 2020-11-18 | | 最終更新日 | 2021-06-02 | | 実験手法 | X-RAY DIFFRACTION (2.083426 Å) | | 主引用文献 | Structural basis for the substrate specificity and catalytic features of pseudouridine kinase from Arabidopsis thaliana.
Nucleic Acids Res., 49, 2021
|
|
6AAE
 
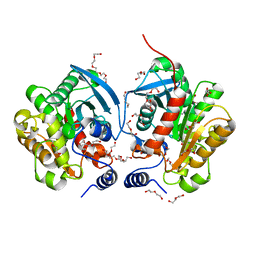 | | Crystal structure of Chloramphenicol-Metabolizaing Enzyme EstDL136 | | 分子名称: | DI(HYDROXYETHYL)ETHER, Esterase, PENTAETHYLENE GLYCOL | | 著者 | Kim, S.H, Kang, P.A, Han, K.T, Lee, S.W, Rhee, S.K. | | 登録日 | 2018-07-18 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.641 Å) | | 主引用文献 | Crystal structure of chloramphenicol-metabolizing enzyme EstDL136 from a metagenome.
PLoS ONE, 14, 2019
|
|
6IEY
 
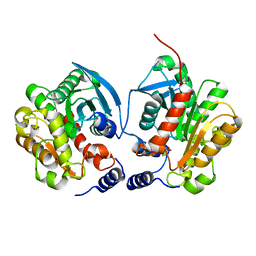 | | Crystal structure of Chloramphenicol-Metabolizaing Enzyme EstDL136-Chloramphenicol complex | | 分子名称: | CHLORAMPHENICOL, Esterase | | 著者 | Kim, S.H, Kang, P.A, Han, K.T, Lee, S.W, Rhee, S.K. | | 登録日 | 2018-09-18 | | 公開日 | 2019-02-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.097 Å) | | 主引用文献 | Crystal structure of chloramphenicol-metabolizing enzyme EstDL136 from a metagenome.
PLoS ONE, 14, 2019
|
|
4UZ0
 
 | | Crystal Structure of apoptosis repressor with CARD (ARC) | | 分子名称: | GLYCEROL, NUCLEOLAR PROTEIN 3 | | 著者 | Kim, S.H, Jeong, J.H, Jang, T.H, Kim, Y.G, Park, H.H. | | 登録日 | 2014-09-04 | | 公開日 | 2015-07-01 | | 最終更新日 | 2017-07-12 | | 実験手法 | X-RAY DIFFRACTION (2.399 Å) | | 主引用文献 | Crystal Structure of Caspase Recruiting Domain (Card) of Apoptosis Repressor with Card (Arc) and its Implication in Inhibition of Apoptosis.
Sci.Rep., 5, 2015
|
|
1HPB
 
 | |
1B8V
 
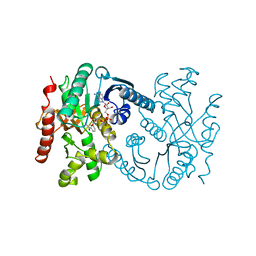 | | Malate dehydrogenase from Aquaspirillum arcticum | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (MALATE DEHYDROGENASE) | | 著者 | Kim, S.Y, Hwang, K.Y, Kim, S.H, Han, Y.S, Cho, Y. | | 登録日 | 1999-02-02 | | 公開日 | 1999-07-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
1EIF
 
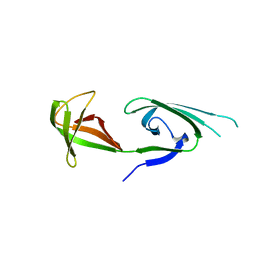 | | EUKARYOTIC TRANSLATION INITIATION FACTOR 5A FROM METHANOCOCCUS JANNASCHII | | 分子名称: | EUKARYOTIC TRANSLATION INITIATION FACTOR 5A | | 著者 | Kim, K.K, Hung, L.W, Yokota, H, Kim, R, Kim, S.H. | | 登録日 | 1998-07-29 | | 公開日 | 1998-10-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1CKP
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR PURVALANOL B | | 分子名称: | 1,2-ETHANEDIOL, PROTEIN (CYCLIN-DEPENDENT PROTEIN KINASE 2), PURVALANOL B | | 著者 | Gray, N.S, Thunnissen, A.M.W.H, Schultz, P.G, Kim, S.H. | | 登録日 | 1998-07-14 | | 公開日 | 1999-01-13 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors.
Science, 281, 1998
|
|
1G8S
 
 | |
1SHS
 
 | |
