7FDA
 
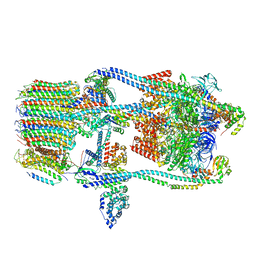 | | CryoEM Structure of Reconstituted V-ATPase, state1 | | Descriptor: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDB
 
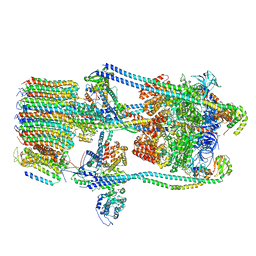 | | CryoEM Structures of Reconstituted V-ATPase,State2 | | Descriptor: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDC
 
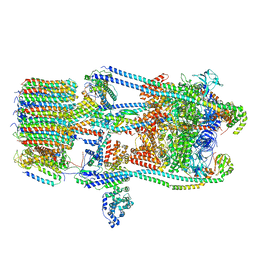 | | CryoEM Structures of Reconstituted V-ATPase, state3 | | Descriptor: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDE
 
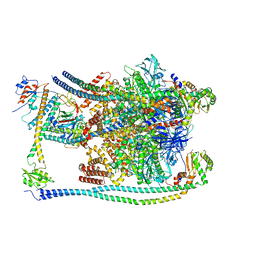 | | CryoEM Structures of Reconstituted V-ATPase, Oxr1 bound V1 | | Descriptor: | Oxidation resistance protein 1, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
2LMA
 
 | | Solution structure of CD4+ T cell derived peptide Thp5 | | Descriptor: | Thp5 peptide | | Authors: | Pandey, N.K, Khan, M.M, Chatterjee, S, Dwivedi, V.P, Tousif, S, Das, J, Van Kaer, L. | | Deposit date: | 2011-11-29 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | CD4+ T cell derived novel peptide Thp5 induces IL-4 production in CD4+ T cells to direct T helper 2 cell differentiation
J.Biol.Chem., 2011
|
|
5JDD
 
 | |
5JDJ
 
 | | Crystal structure of domain I10 from titin in space group P212121 | | Descriptor: | CALCIUM ION, Titin | | Authors: | Williams, R, Bogomolovas, J, Labiet, S, Mayans, O. | | Deposit date: | 2016-04-16 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Exploration of pathomechanisms triggered by a single-nucleotide polymorphism in titin's I-band: the cardiomyopathy-linked mutation T2580I.
Open Biology, 6, 2016
|
|
4WNI
 
 | |
4WNC
 
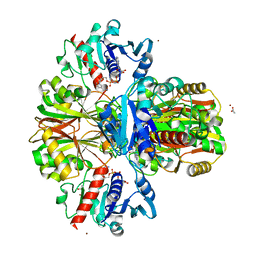 | | Crystal structure of human wild-type GAPDH at 1.99 angstroms resolution | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Garcin, E.D. | | Deposit date: | 2014-10-11 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A Dimer Interface Mutation in Glyceraldehyde-3-Phosphate Dehydrogenase Regulates Its Binding to AU-rich RNA.
J.Biol.Chem., 290, 2015
|
|
5JDE
 
 | |
4M3L
 
 | | Crystal Structure of the coiled coil domain of MuRF1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, E3 ubiquitin-protein ligase TRIM63, ... | | Authors: | Mayans, O, Franke, B. | | Deposit date: | 2013-08-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the fold organization and sarcomeric targeting of the muscle atrogin MuRF1.
Open Biol, 4, 2014
|
|
4QEG
 
 | |
