7D6P
 
 | |
7D6T
 
 | |
4KRU
 
 | |
4KRT
 
 | |
7F5I
 
 | | X-ray structure of Clostridium perfringens-specific amidase endolysin | | Descriptor: | GLUTAMIC ACID, SODIUM ION, ZINC ION, ... | | Authors: | Kamitori, S, Tamai, E. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biochemical characterization of the Clostridium perfringens-specific Zn 2+ -dependent amidase endolysin, Psa, catalytic domain.
Biochem.Biophys.Res.Commun., 576, 2021
|
|
7CFL
 
 | |
5XCC
 
 | |
5XCB
 
 | |
6L68
 
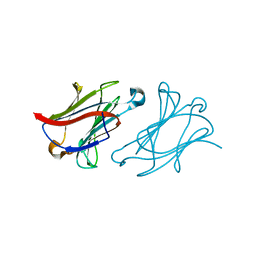 | | X-ray structure of human galectin-10 in complex with D-allose | | Descriptor: | Galectin-10, beta-D-allopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
6L67
 
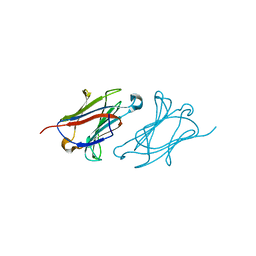 | | X-ray structure of human galectin-10 in complex with D-galactose | | Descriptor: | Galectin-10, beta-D-galactopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
6L6D
 
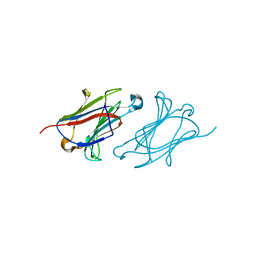 | | X-ray structure of human galectin-10 in complex with D-N-acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Galectin-10 | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
6L6B
 
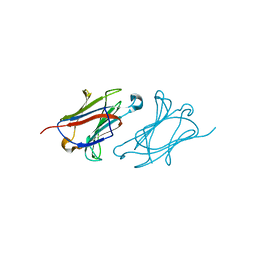 | | X-ray structure of human galectin-10 in complex with L-fucose | | Descriptor: | Galectin-10, beta-L-fucopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
6L6C
 
 | | X-ray structure of human galectin-10 in complex with D-arabinose | | Descriptor: | Galectin-10, alpha-D-arabinopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
6L64
 
 | | X-ray structure of human galectin-10 in complex with D-glucose | | Descriptor: | Galectin-10, beta-D-glucopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
6L6A
 
 | | X-ray structure of human galectin-10 in complex with D-mannose | | Descriptor: | Galectin-10, beta-D-mannopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
3VKN
 
 | | Galectin-8 N-terminal domain in free form | | Descriptor: | CHLORIDE ION, Galectin-8 | | Authors: | Kamitori, S, Yoshida, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains
Febs J., 279, 2012
|
|
3VKL
 
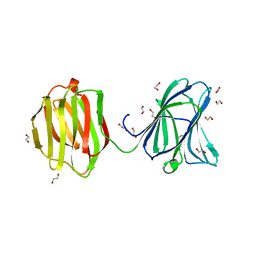 | |
3VKO
 
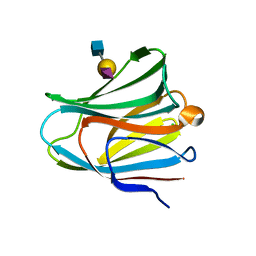 | | Galectin-8 N-terminal domain in complex with sialyllactosamine | | Descriptor: | CHLORIDE ION, Galectin-8, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kamitori, S, Yoshida, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains
Febs J., 279, 2012
|
|
5YFK
 
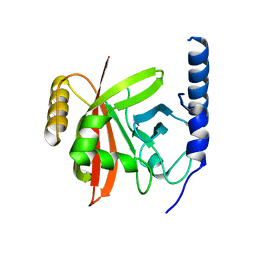 | |
2D58
 
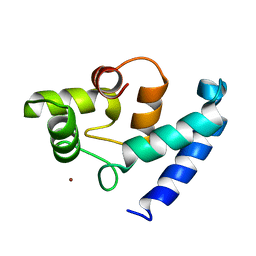 | | Human microglia-specific protein Iba1 | | Descriptor: | Allograft inflammatory factor 1, NICKEL (II) ION | | Authors: | Kamitori, S. | | Deposit date: | 2005-10-31 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Structures of the Microglia/Macrophage-specific Protein Iba1 from Human and Mouse Demonstrate Novel Molecular Conformation Change Induced by Calcium binding
J.Mol.Biol., 364, 2006
|
|
3B4X
 
 | |
5J8L
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy L-tagatose, using a crystal grown in microgravity | | Descriptor: | 1-deoxy-L-tagatose, 1-deoxy-beta-L-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Izumori, K, Kamitori, S. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
5X2J
 
 | | Crystal structure of a recombinant hybrid manganese superoxide dismutase from Staphylococcus equorum and Staphylococcus saprophyticus | | Descriptor: | MANGANESE (II) ION, manganese superoxide dismutase | | Authors: | Retnoningrum, D.S, Yoshida, H, Arumsari, S, Kamitori, S, Ismaya, W.T. | | Deposit date: | 2017-01-31 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The first crystal structure of manganese superoxide dismutase from the genus Staphylococcus
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3NV2
 
 | |
3NV1
 
 | | Crystal structure of human galectin-9 C-terminal CRD | | Descriptor: | Galectin 9 short isoform variant, NICKEL (II) ION | | Authors: | Yoshida, H, Kamitori, S. | | Deposit date: | 2010-07-07 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structures of human galectin-9 C-terminal domain in complexes with a biantennary oligosaccharide and sialyllactose
J.Biol.Chem., 285, 2010
|
|
