1OXU
 
 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Verdon, G, Albers, S.V, Dijkstra, B.W, Driessen, A.J, Thunnissen, A.M. | | Deposit date: | 2003-04-03 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus: nucleotide-free and nucleotide-bound conformations
J.Mol.Biol., 330, 2003
|
|
1IIB
 
 | | CRYSTAL STRUCTURE OF IIBCELLOBIOSE FROM ESCHERICHIA COLI | | Descriptor: | ENZYME IIB OF THE CELLOBIOSE-SPECIFIC PHOSPHOTRANSFERASE SYSTEM | | Authors: | Van Montfort, R.L.M, Pijning, T, Kalk, K.H, Reizer, J, Saier, M.H, Thunnissen, M.M.G.M, Robillard, G.T, Dijkstra, B.W. | | Deposit date: | 1996-12-23 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of an energy-coupling protein from bacteria, IIBcellobiose, reveals similarity to eukaryotic protein tyrosine phosphatases.
Structure, 5, 1997
|
|
1JUH
 
 | | Crystal Structure of Quercetin 2,3-dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fusetti, F, Schroeter, K.H, Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2001-08-24 | | Release date: | 2002-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the copper-containing quercetin 2,3-dioxygenase from Aspergillus japonicus.
Structure, 10, 2002
|
|
1JX9
 
 | | Penicillin Acylase, mutant | | Descriptor: | CALCIUM ION, penicillin G acylase alpha subunit, penicillin G acylase beta subunit | | Authors: | Hensgens, C.M.H, Keizer, E, Snijder, H.J, Dijkstra, B.W. | | Deposit date: | 2001-09-06 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural and kinetic studies on ligand binding in wild-type and active-site mutants of penicillin acylase.
Protein Eng.Des.Sel., 17, 2004
|
|
1K7D
 
 | | Penicillin Acylase with Phenyl Proprionic Acid | | Descriptor: | CALCIUM ION, Penicillin Acylase alpha subunit, R-2-PHENYL-PROPRIONIC ACID, ... | | Authors: | Hensgens, C.M.H, Keizer, E, Snijder, H.J, Dijkstra, B.W. | | Deposit date: | 2001-10-19 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and kinetic studies on ligand binding in wild-type and active-site mutants of penicillin acylase.
Protein Eng.Des.Sel., 17, 2004
|
|
1K5Q
 
 | | PENICILLIN ACYLASE, MUTANT COMPLEXED WITH PAA | | Descriptor: | 2-PHENYLACETIC ACID, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | Hensgens, C.M.H, Keizer, E, Snijder, H.J, Dijkstra, B.W. | | Deposit date: | 2001-10-12 | | Release date: | 2003-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural and kinetic studies on ligand binding in wild-type and active-site mutants of penicillin acylase.
Protein Eng.Des.Sel., 17, 2004
|
|
1K5S
 
 | | PENICILLIN ACYLASE, MUTANT COMPLEXED WITH PPA | | Descriptor: | CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, PENICILLIN G ACYLASE BETA SUBUNIT, ... | | Authors: | Hensgens, C.M.H, Keizer, E, Snijder, H.J, Dijkstra, B.W. | | Deposit date: | 2001-10-12 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and kinetic studies on ligand binding in wild-type and active-site mutants of penicillin acylase.
Protein Eng.Des.Sel., 17, 2004
|
|
1KCK
 
 | | Bacillus circulans strain 251 Cyclodextrin glycosyl transferase mutant N193G | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Rozeboom, H.J, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 2001-11-09 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The remote substrate binding subsite -6 in cyclodextrin-glycosyltransferase controls the transferase activity of the enzyme via an induced-fit mechanism.
J.Biol.Chem., 277, 2002
|
|
1KCL
 
 | | Bacillus ciruclans strain 251 Cyclodextrin glycosyl transferase mutant G179L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Cyclodextrin glycosyltransferase, ... | | Authors: | Rozeboom, H.J, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 2001-11-09 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The remote substrate binding subsite -6 in cyclodextrin-glycosyltransferase controls the transferase activity of the enzyme via an induced-fit mechanism.
J.Biol.Chem., 277, 2002
|
|
1KEC
 
 | | PENICILLIN ACYLASE MUTANT WITH PHENYL PROPRIONIC ACID | | Descriptor: | CALCIUM ION, PENICILLIN ACYLASE ALPHA SUBUNIT, PENICILLIN ACYLASE BETA SUBUNIT, ... | | Authors: | Hensgens, C.M.H, Keizer, E, Snijder, H.J, Dijkstra, B.W. | | Deposit date: | 2001-11-15 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic studies on ligand binding in wild-type and active-site mutants of penicillin acylase.
Protein Eng.Des.Sel., 17, 2004
|
|
1AQ6
 
 | | STRUCTURE OF L-2-HALOACID DEHALOGENASE FROM XANTHOBACTER AUTOTROPHICUS | | Descriptor: | FORMIC ACID, L-2-HALOACID DEHALOGENASE | | Authors: | Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1997-08-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-dimensional structure of L-2-haloacid dehalogenase from Xanthobacter autotrophicus GJ10 complexed with the substrate-analogue formate.
J.Biol.Chem., 272, 1997
|
|
1C9U
 
 | | CRYSTAL STRUCTURE OF THE SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE IN COMPLEX WITH PQQ | | Descriptor: | CALCIUM ION, GLYCEROL, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-08-03 | | Release date: | 2000-02-04 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of soluble quinoprotein glucose dehydrogenase.
EMBO J., 18, 1999
|
|
1CDG
 
 | | NUCLEOTIDE SEQUENCE AND X-RAY STRUCTURE OF CYCLODEXTRIN GLYCOSYLTRANSFERASE FROM BACILLUS CIRCULANS STRAIN 251 IN A MALTOSE-DEPENDENT CRYSTAL FORM | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYL-TRANSFERASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Lawson, C.L, Van Montfort, R, Strokopytov, B.V, Kalk, K.H, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1993-08-02 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nucleotide sequence and X-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 in a maltose-dependent crystal form.
J.Mol.Biol., 236, 1994
|
|
1CQ1
 
 | | Soluble Quinoprotein Glucose Dehydrogenase from Acinetobacter Calcoaceticus in Complex with PQQH2 and Glucose | | Descriptor: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-08-04 | | Release date: | 2000-02-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of soluble quinoprotein glucose dehydrogenase.
EMBO J., 18, 1999
|
|
1EHY
 
 | | X-ray structure of the epoxide hydrolase from agrobacterium radiobacter ad1 | | Descriptor: | POTASSIUM ION, PROTEIN (SOLUBLE EPOXIDE HYDROLASE) | | Authors: | Nardini, M, Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Rink, R, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 1998-10-17 | | Release date: | 1999-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The x-ray structure of epoxide hydrolase from Agrobacterium radiobacter AD1. An enzyme to detoxify harmful epoxides.
J.Biol.Chem., 274, 1999
|
|
1LG1
 
 | |
1LG2
 
 | |
1LTM
 
 | | ACCELERATED X-RAY STRUCTURE ELUCIDATION OF A 36 KDA MURAMIDASE/TRANSGLYCOSYLASE USING WARP | | Descriptor: | 1,2-ETHANEDIOL, 36 KDA SOLUBLE LYTIC TRANSGLYCOSYLASE, BICINE, ... | | Authors: | Van Asselt, E.J, Dijkstra, B.W. | | Deposit date: | 1997-09-26 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accelerated X-ray structure elucidation of a 36 kDa muramidase/transglycosylase using wARP.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1LQ0
 
 | |
1MPX
 
 | | ALPHA-AMINO ACID ESTER HYDROLASE LABELED WITH SELENOMETHIONINE | | Descriptor: | CALCIUM ION, GLYCEROL, alpha-amino acid ester hydrolase | | Authors: | Barends, T.R.M, Polderman-Tijmes, J.J, Jekel, P.A, Hensgens, C.M.H, de Vries, E.J, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2002-09-13 | | Release date: | 2003-04-15 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The sequence and crystal structure of the alpha-amino acid ester hydrolase from Xanthomonas citri define a new family of beta-lactam antibiotic acylases.
J.Biol.Chem., 278, 2003
|
|
1NHC
 
 | | Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | van Pouderoyen, G, Snijder, H.J, Benen, J.A, Dijkstra, B.W. | | Deposit date: | 2002-12-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger.
Febs Lett., 554, 2003
|
|
1NWU
 
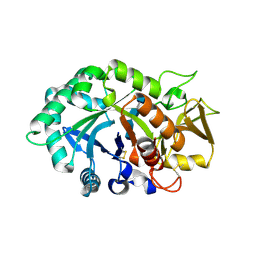 | | Crystal structure of human cartilage gp39 (HC-gp39) in complex with chitotetraose | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fusetti, F, Pijning, T, Kalk, K.H, Bos, E, Dijkstra, B.W. | | Deposit date: | 2003-02-06 | | Release date: | 2003-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and carbohydrate-binding properties of the human cartilage glycoprotein-39
J.Biol.Chem., 278, 2003
|
|
1NWS
 
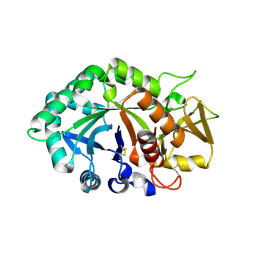 | | Crystal structure of human cartilage gp39 (HC-gp39) in complex with chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3 like protein 1 | | Authors: | Fusetti, F, Pijning, T, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2003-02-06 | | Release date: | 2003-08-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and carbohydrate-binding properties of the human cartilage glycoprotein-39
J.Biol.Chem., 278, 2003
|
|
1NX9
 
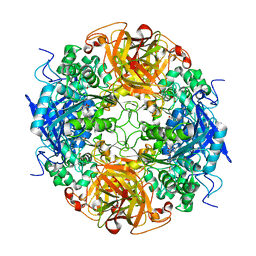 | | Acetobacter turbidans alpha-amino acid ester hydrolase S205A mutant complexed with ampicillin | | Descriptor: | (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, GLYCEROL, alpha-amino acid ester hydrolase | | Authors: | Barends, T.R.M, Polderman-Tijmes, J.J, Jekel, P.A, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2003-02-10 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acetobacter turbidans {alpha}-Amino Acid Ester Hydrolase: HOW A SINGLE MUTATION IMPROVES AN ANTIBIOTIC-PRODUCING ENZYME.
J.Biol.Chem., 281, 2006
|
|
1NWR
 
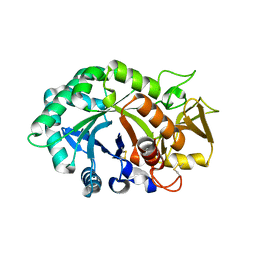 | | Crystal structure of human cartilage gp39 (HC-gp39) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3 like protein 1 | | Authors: | Fusetti, F, Pijning, T, Kalk, K.H, Bos, E, Dijkstra, B.W. | | Deposit date: | 2003-02-06 | | Release date: | 2003-08-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and Carbohydrate-binding Properties of the Human Cartilage Glycoprotein-39
J.Biol.Chem., 278, 2003
|
|
