3JAU
 
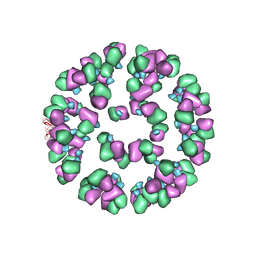 | | The cryoEM map of EV71 mature viron in complex with the Fab fragment of antibody D5 | | Descriptor: | Capsid protein VP1, Heavy chain of Fab fragment variable region of antibody D5, Light chain of Fab fragment variable region of antibody D5 | | Authors: | Fan, C, Ye, X.H, Ku, Z.Q, Zuo, T, Kong, L.L, Zhang, C, Shi, J.P, Liu, Q.W, Chen, T, Zhang, Y.Y, Jiang, W, Zhang, L.Q, Huang, Z, Cong, Y. | | Deposit date: | 2015-06-24 | | Release date: | 2016-02-10 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural Basis for Recognition of Human Enterovirus 71 by a Bivalent Broadly Neutralizing Monoclonal Antibody
Plos Pathog., 12, 2016
|
|
5TSL
 
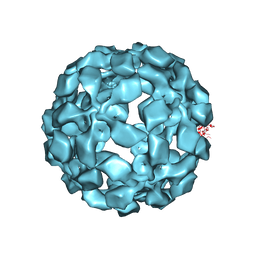 | |
5TSK
 
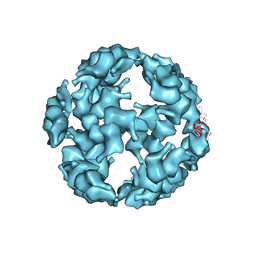 | |
6KRE
 
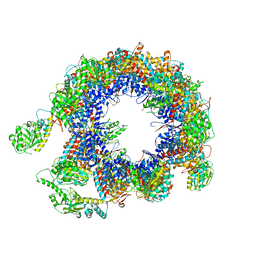 | | TRiC at 0.05 mM ADP-AlFx, Conformation 2, 0.05-C2 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KRD
 
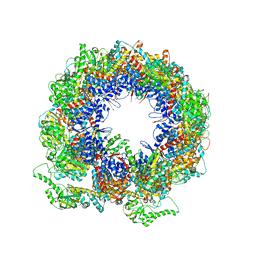 | | TRiC at 0.05 mM ADP-AlFx, Conformation 4, 0.05-C4 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KS8
 
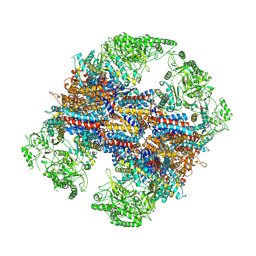 | | TRiC at 0.1 mM ADP-AlFx, Conformation 4, 0.1-C4 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KS7
 
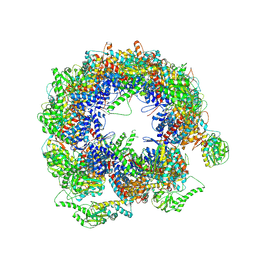 | | TRiC at 0.1 mM ADP-AlFx, Conformation 1, 0.1-C1 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.62 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KS6
 
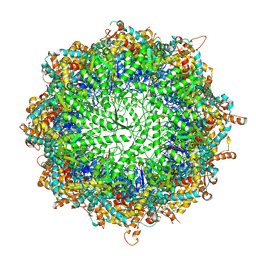 | | TRiC at 0.2 mM ADP-AlFx, Conformation 1, 0.2-C1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IIJ
 
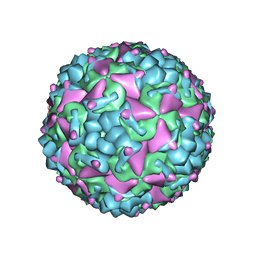 | | Cryo-EM structure of CV-A10 mature virion | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Chen, J.H, Ye, X.H, Cong, Y, Huang, Z. | | Deposit date: | 2018-10-06 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Coxsackievirus A10 atomic structure facilitating the discovery of a broad-spectrum inhibitor against human enteroviruses.
Cell Discov, 5, 2019
|
|
6IIO
 
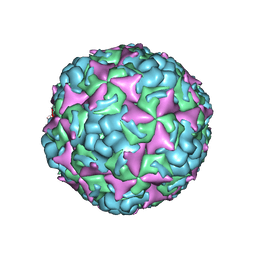 | | Cryo-EM structure of CV-A10 native empty particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Chen, J.H, Ye, X.H, Cong, Y, Huang, Z. | | Deposit date: | 2018-10-07 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Coxsackievirus A10 atomic structure facilitating the discovery of a broad-spectrum inhibitor against human enteroviruses.
Cell Discov, 5, 2019
|
|
5WVK
 
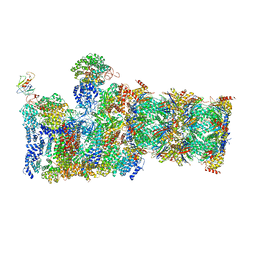 | | Yeast proteasome-ADP-AlFx | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
5YHQ
 
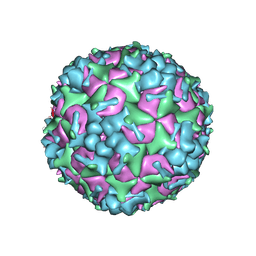 | | Cryo-EM Structure of CVA6 VLP | | Descriptor: | Capsid protein VP1, Capsid protein VP3, capsid protein VP0 | | Authors: | Chen, J, Zhang, C, Huang, Z, Cong, Y. | | Deposit date: | 2017-09-29 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 3.0-Angstrom Resolution Cryo-Electron Microscopy Structure and Antigenic Sites of Coxsackievirus A6-Like Particles.
J. Virol., 92, 2018
|
|
7FIF
 
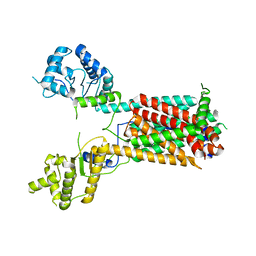 | | Cryo-EM structure of the hedgehog release protein Disp from water bear (Hypsibius dujardini) | | Descriptor: | Protein dispatched-like protein 1 | | Authors: | Luo, Y, Wan, G, Wang, Q, Zhao, Y, Cong, Y, Li, D. | | Deposit date: | 2021-07-31 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Architecture of Dispatched, a Transmembrane Protein Responsible for Hedgehog Release.
Front Mol Biosci, 8, 2021
|
|
7DK3
 
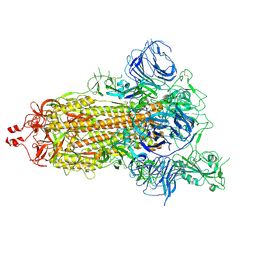 | | SARS-CoV-2 S trimer, S-open | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-16 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7DR7
 
 | | bovine 20S immunoproteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2020-12-26 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM of mammalian PA28 alpha beta-iCP immunoproteasome reveals a distinct mechanism of proteasome activation by PA28 alpha beta.
Nat Commun, 12, 2021
|
|
7EKO
 
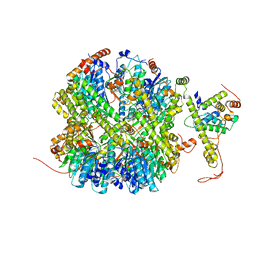 | | CrClpP-S1 | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit | | Authors: | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
7EKQ
 
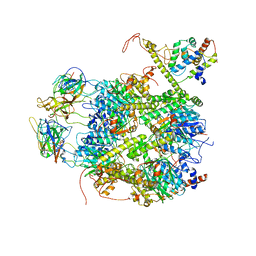 | | CrClpP-S2c | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
7DMP
 
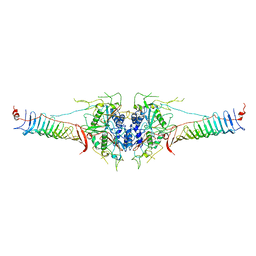 | | Mouse radial spoke complex | | Descriptor: | Radial spoke head 1 homolog, Radial spoke head protein 4 homolog A, Radial spoke head protein 9 homolog | | Authors: | Zheng, W, Cong, Y. | | Deposit date: | 2020-12-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Distinct architecture and composition of mouse axonemal radial spoke head revealed by cryo-EM
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EBZ
 
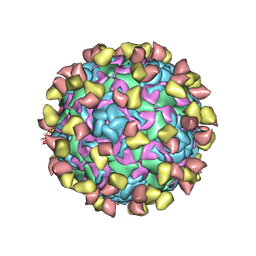 | | EV-D68 in complex with 2H12 Fab (state S1) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
7EBR
 
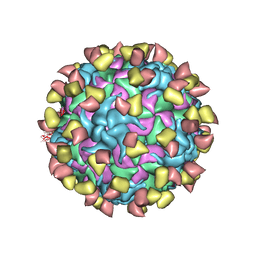 | | EV-D68 in complex with 2H12 Fab (state S2) | | Descriptor: | 2H12 Fab heavy chain, 2H12 Fab light chain, Capsid protein VP1, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
7EC5
 
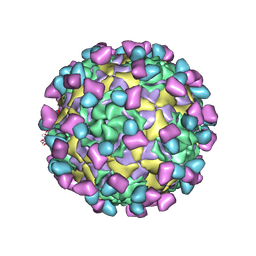 | | EV-D68 in complex with 8F12 Fab | | Descriptor: | 8F12 Fab heavy chain, 8F12 Fab light chain, Capsid protein VP1, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
7ECY
 
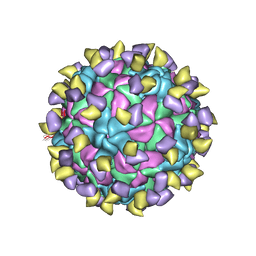 | | EV-D68 in complex with 2H12 Fab (State 3) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
7WD8
 
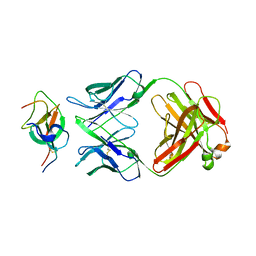 | | SARS-CoV-2 Beta spike SD1 in complex with S3H3 Fab | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD9
 
 | |
7WD7
 
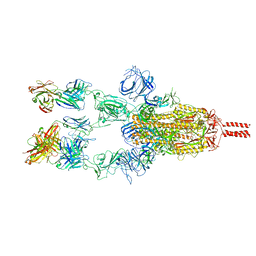 | |
