3GFO
 
 | | Structure of cbiO1 from clostridium perfringens: Part of the ABC transporter complex cbiONQ. | | 分子名称: | Cobalt import ATP-binding protein cbiO 1, SULFATE ION | | 著者 | Ramagopal, U.A, Morano, C, Toro, R, Dickey, M, Do, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-02-27 | | 公開日 | 2009-03-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of cbiO1 from clostridium perfringens: Part of the ABC transporter complex cbiONQ
To be published
|
|
1J2X
 
 | | Crystal structure of RAP74 C-terminal domain complexed with FCP1 C-terminal peptide | | 分子名称: | RNA polymerase II CTD phosphatase, SULFATE ION, Transcription initiation factor IIF, ... | | 著者 | Kamada, K, Roeder, R.G, Burley, S.K. | | 登録日 | 2003-01-15 | | 公開日 | 2003-01-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular mechanism of recruitment of TFIIF- associating RNA polymerase C-terminal domain phosphatase (FCP1) by transcription factor IIF
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NKP
 
 | | Crystal structure of Myc-Max recognizing DNA | | 分子名称: | 5'-D(*CP*GP*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*TP*C)-3', Max protein, Myc proto-oncogene protein | | 著者 | Nair, S.K, Burley, S.K. | | 登録日 | 2003-01-03 | | 公開日 | 2003-02-04 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray structures of Myc-Max and Mad-Max recognizing DNA: Molecular bases of regulation by proto-oncogenic transcription factors
Cell(Cambridge,Mass.), 112, 2003
|
|
1NLW
 
 | | Crystal structure of Mad-Max recognizing DNA | | 分子名称: | 5'-D(*GP*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*TP*C)-3', MAD PROTEIN, MAX PROTEIN | | 著者 | Nair, S.K, Burley, S.K. | | 登録日 | 2003-01-07 | | 公開日 | 2003-02-04 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray structures of Myc-Max and Mad-Max recognizing DNA: Molecular bases of regulation by proto-oncogenic transcription factors
Cell(Cambridge,Mass.), 112, 2003
|
|
4F0R
 
 | | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) bound Zn and 5'-Methylthioadenosine (unproductive complex) | | 分子名称: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5-methylthioadenosine/S-adenosylhomocysteine deaminase, GLYCEROL, ... | | 著者 | Kim, J, Vetting, M.W, Sauder, J.M, Burley, S.K, Raushel, F.M, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-05-04 | | 公開日 | 2012-06-06 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) bound Zn and 5'-Methylthioadenosine (unproductive complex)
To be Published
|
|
4F2D
 
 | | Crystal Structure of Escherichia coli L-arabinose Isomerase (ECAI) complexed with Ribitol | | 分子名称: | ACETIC ACID, D-ribitol, L-arabinose isomerase, ... | | 著者 | Manjasetty, B.A, Burley, S.K, Almo, S.C, Chance, M.R, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2012-05-07 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of Escherichia coli L-arabinose Isomerase (ECAI) complexed with Ribitol
TO BE PUBLISHED
|
|
4F0S
 
 | | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) with bound inosine. | | 分子名称: | 5-methylthioadenosine/S-adenosylhomocysteine deaminase, CHLORIDE ION, INOSINE, ... | | 著者 | Kim, J, Vetting, M.W, Sauder, J.M, Burley, S.K, Raushel, F.M, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-05-04 | | 公開日 | 2012-06-06 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.851 Å) | | 主引用文献 | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) with bound inosine.
To be Published
|
|
4HL7
 
 | | Crystal structure of nicotinate phosphoribosyltransferase (target NYSGR-026035) from Vibrio cholerae | | 分子名称: | Nicotinate phosphoribosyltransferase, SULFATE ION | | 著者 | Mulichak, A.M, Sauder, J.M, Keefe, L.J, Burley, S.K, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-10-16 | | 公開日 | 2012-11-14 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of nicotinate phosphoribosyltransferase (target NYSGR-026035) from Vibrio cholerae
To be Published
|
|
4HYR
 
 | | Structure of putative Glucarate dehydratase from Acidaminococcus sp. D21 with unusual static disorder | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Hegde, R.P, Toro, R, Burley, S.K, Almo, S.C, Ramagopal, U.A, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-11-14 | | 公開日 | 2013-02-13 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structure of putative Glucarate dehydratase from Acidaminococcus sp. D21 with unusual static disorder
To be published
|
|
4HN8
 
 | | Crystal structure of a putative D-glucarate dehydratase from Pseudomonas mendocina ymp | | 分子名称: | D-glucarate dehydratase, GLYCEROL | | 著者 | Hegde, R.P, Toro, R, Burley, S.K, Almo, S.C, Ramagopal, U.A, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-10-19 | | 公開日 | 2012-11-07 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a putative D-glucarate dehydratase from Pseudomonas mendocina ymp
To be published
|
|
5QC0
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | 分子名称: | 2-(dimethylamino)-1-[4-(2-oxo-2-{3-[3-{[2-(piperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-1-propyl-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethyl)piperidin-1-yl]ethan-1-one, Cathepsin S | | 著者 | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | 登録日 | 2017-08-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCG
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | 分子名称: | Cathepsin S, N-benzyl-1-{2-chloro-5-[2-(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethyl]phenyl}methanamine, SULFATE ION | | 著者 | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | 登録日 | 2017-08-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCY
 
 | | Crystal structure of BACE complex with BMC008 | | 分子名称: | (9R,11S)-11-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-9-methyl-16-(1,3-oxazol-2-yl)-3-[(1R)-1-phenylethyl]-3,12-diazabicyclo[12.3.1]octadeca-1(18),14,16-triene-2,13-dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QC9
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | 分子名称: | Cathepsin S, N-{[5-{1-[3-(dimethylamino)propyl]-5-[(piperidin-4-yl)acetyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}-2-(trifluoromethyl)phenyl]methyl}-3-methylbut-2-enamide | | 著者 | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | 登録日 | 2017-08-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCP
 
 | | Crystal structure of BACE complex with BMC018 | | 分子名称: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-19-(2-oxopropoxy)-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCD
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | 分子名称: | Cathepsin S, SULFATE ION, {(3S)-7-[(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]-1,2,3,4-tetrahydroisoquinolin-3-yl}(piperidin-1-yl)methanone | | 著者 | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | 登録日 | 2017-08-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QD5
 
 | | Crystal structure of BACE complex with BMC009 | | 分子名称: | (10S,12S)-17-chloro-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-10-methyl-7-oxa-2,13,18-triazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCX
 
 | | Crystal structure of BACE complex with BMC007 | | 分子名称: | (9R,11S)-3-ethyl-11-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-9-methyl-3,12-diazabicyclo[12.3.1]octadeca-1(18),14,16-triene-2,13-dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD9
 
 | | Crystal structure of BACE complex with BMC005 | | 分子名称: | (5S,8S,10R)-8-[(1R)-2-{[1-(3-tert-butylphenyl)cyclopropyl]amino}-1-hydroxyethyl]-4,5,10-trimethyl-1-oxa-4,7-diazacyclohexadecane-3,6-dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.602 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QC2
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | 分子名称: | 2-[1-(cyclohexylmethyl)piperidin-4-yl]-1-{3-[3-{[2-(4-fluoropiperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-1-(3-hydroxypropyl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, Cathepsin S | | 著者 | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | 登録日 | 2017-08-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCI
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | 分子名称: | Cathepsin S, GLYCEROL, N-benzyl-1-{5-[(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]-2-methoxyphenyl}methanamine, ... | | 著者 | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | 登録日 | 2017-08-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.179 Å) | | 主引用文献 | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCU
 
 | | Crystal structure of BACE complex with BMC022 | | 分子名称: | (2R,4S)-N-butyl-4-[(5S,8S,10R)-5,10-dimethyl-3,3,6-trioxo-3lambda~6~-thia-7-azabicyclo[11.3.1]heptadeca-1(17),13,15-trien-8-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD6
 
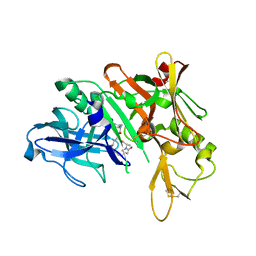 | | Crystal structure of BACE complex with BMC004 | | 分子名称: | (3S,14R,16S)-16-[1,1-dihydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-3,4,14-trimethyl-1,4-diazacyclohexadecane-2,5-dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QBX
 
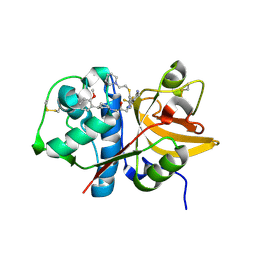 | | Crystal structure of human Cathepsin-S with bound ligand | | 分子名称: | (2S)-1-[4-(2-methoxyphenyl)piperidin-1-yl]-3-{3-[3-{[2-(piperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-1-yl}propan-2-ol, Cathepsin S | | 著者 | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | 登録日 | 2017-08-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCE
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | 分子名称: | Cathepsin S, N-benzyl-1-{2-chloro-5-[(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]phenyl}methanamine | | 著者 | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | 登録日 | 2017-08-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
