2VLA
 
 | | Crystal structure of restriction endonuclease BpuJI recognition domain in complex with cognate DNA | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 5'-D(*GP*GP*TP*AP*CP*CP*CP*GP*TP*GP *GP*A)-3', 5'-D(*TP*CP*CP*AP*CP*GP*GP*GP*TP*AP *CP*C)-3', ... | | Authors: | Sukackaite, R, Grazulis, S, Bochtler, M, Siksnys, V. | | Deposit date: | 2008-01-11 | | Release date: | 2008-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Recognition Domain of the Bpuji Restriction Endonuclease in Complex with Cognate DNA at 1.3-A Resolution.
J.Mol.Biol., 378, 2008
|
|
3PRH
 
 | | tryptophanyl-tRNA synthetase Val144Pro mutant from B. subtilis | | Descriptor: | Tryptophanyl-tRNA synthetase | | Authors: | Antonczak, A.K, Simova, Z, Yonemoto, I, Bochtler, M, Piasecka, A, Czapinska, H, Brancale, A, Tippmann, E.M. | | Deposit date: | 2010-11-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Importance of single molecular determinants in the fidelity of expanded genetic codes.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2B0P
 
 | | truncated S. aureus LytM, P212121 crystal form | | Descriptor: | ACETATE ION, CACODYLATE ION, Glycyl-glycine endopeptidase lytM, ... | | Authors: | Firczuk, M, Mucha, A, Bochtler, M. | | Deposit date: | 2005-09-14 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of active LytM.
J.Mol.Biol., 354, 2005
|
|
2B13
 
 | | Truncated S. aureus LytM, P41 crystal form | | Descriptor: | Glycyl-glycine endopeptidase lytM, L(+)-TARTARIC ACID, ZINC ION | | Authors: | Firczuk, M, Mucha, A, Bochtler, M. | | Deposit date: | 2005-09-15 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of active LytM.
J.Mol.Biol., 354, 2005
|
|
1X9Y
 
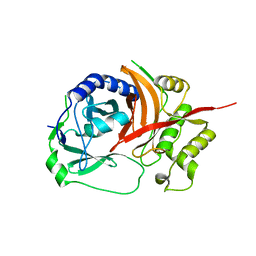 | | The prostaphopain B structure | | Descriptor: | cysteine proteinase | | Authors: | Filipek, R, Szczepanowski, R, Sabat, A, Potempa, J, Bochtler, M. | | Deposit date: | 2004-08-24 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Prostaphopain B structure: a comparison of proregion-mediated and staphostatin-mediated protease inhibition.
Biochemistry, 43, 2004
|
|
2B44
 
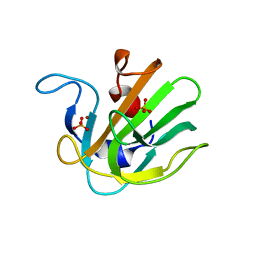 | | Truncated S. aureus LytM, P 32 2 1 crystal form | | Descriptor: | Glycyl-glycine endopeptidase lytM, PHOSPHATE ION, ZINC ION | | Authors: | Firczuk, M, Mucha, A, Bochtler, M. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of active LytM.
J.Mol.Biol., 354, 2005
|
|
1YR3
 
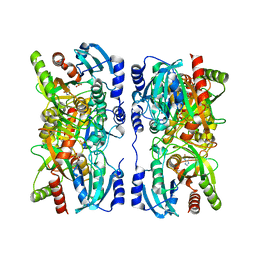 | | Escherichia coli purine nucleoside phosphorylase II, the product of the xapA gene | | Descriptor: | SULFATE ION, XANTHINE, Xanthosine phosphorylase | | Authors: | Dandanell, G, Szczepanowski, R.H, Kierdaszuk, B, Shugar, D, Bochtler, M. | | Deposit date: | 2005-02-03 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Escherichia coli purine nucleoside phosphorylase II, the product of the xapA gene
J.Mol.Biol., 348, 2005
|
|
1YQU
 
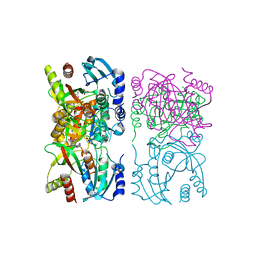 | | Escherichia coli purine nucleoside phosphorylase II, the product of the xapA gene | | Descriptor: | GUANINE, PHOSPHATE ION, Xanthosine phosphorylase | | Authors: | Dandanell, G, Szczepanowski, R.H, Kierdaszuk, B, Shugar, D, Bochtler, M. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Escherichia coli purine nucleoside phosphorylase II, the product of the xapA gene
J.Mol.Biol., 348, 2005
|
|
1YQQ
 
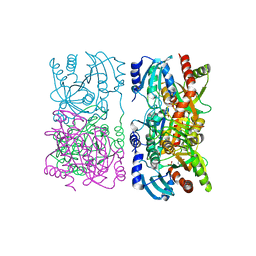 | | Escherichia coli purine nucleoside phosphorylase II, the product of the xapA gene | | Descriptor: | GUANINE, PHOSPHATE ION, Xanthosine phosphorylase | | Authors: | Dandanell, G, Szczepanowski, R.H, Kierdaszuk, B, Shugar, D, Bochtler, M. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Escherichia coli Purine Nucleoside Phosphorylase II, the Product of the xapA Gene
J.Mol.Biol., 348, 2005
|
|
1Z7L
 
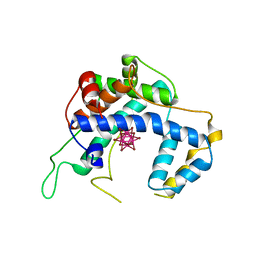 | |
1ZJC
 
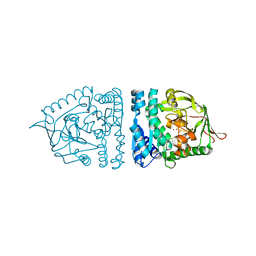 | | Aminopeptidase S from S. aureus | | Descriptor: | COBALT (II) ION, aminopeptidase ampS | | Authors: | Odintsov, S.G, Sabala, I, Bourenkov, G, Rybin, V, Bochtler, M. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Staphylococcus aureus Aminopeptidase S Is a Founding Member of a New Peptidase Clan.
J.Biol.Chem., 280, 2005
|
|
1ZRS
 
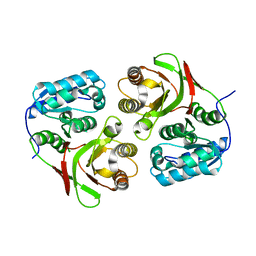 | | wild-type LD-carboxypeptidase | | Descriptor: | hypothetical protein | | Authors: | Korza, H.J, Bochtler, M. | | Deposit date: | 2005-05-21 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pseudomonas aeruginosa LD-carboxypeptidase, a serine peptidase with a Ser-His-Glu triad and a nucleophilic elbow.
J.Biol.Chem., 280, 2005
|
|
2AYI
 
 | | Wild-type AmpT from Thermus thermophilus | | Descriptor: | Aminopeptidase T, ZINC ION | | Authors: | Odintsov, S.G, Sabala, I, Bourenkov, G, Rybin, V, Bochtler, M. | | Deposit date: | 2005-09-07 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Substrate Access to the Active Sites in Aminopeptidase T, a Representative of a New Metallopeptidase Clan.
J.Mol.Biol., 354, 2005
|
|
2AUN
 
 | |
2AUM
 
 | |
1M4Y
 
 | | Crystal structure of HslV from Thermotoga maritima | | Descriptor: | ATP-dependent protease hslV, SODIUM ION | | Authors: | Song, H.K, Ramachandran, R, Bochtler, M.B, Hartmann, C, Azim, M.K, Huber, R. | | Deposit date: | 2002-07-05 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isolation and characterization of the prokaryotic proteasome homolog HslVU (ClpQY) from Thermotoga maritima and the crystal structure of HslV.
BIOPHYS.CHEM., 100, 2003
|
|
8UOO
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum | | Descriptor: | Asparaginase, CHLORIDE ION, GLYCEROL | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UOW
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21A) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Asparaginase | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP6
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K19A) in complex with L-Asp | | Descriptor: | ASPARTIC ACID, Asparaginase, TETRAETHYLENE GLYCOL | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UOR
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K19E) | | Descriptor: | 1,2-ETHANEDIOL, Asparaginase, CHLORIDE ION | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP8
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21F, complex with L-Asp) | | Descriptor: | ASPARTIC ACID, Asparaginase, CHLORIDE ION, ... | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UOU
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum in complex with L-Asp | | Descriptor: | ASPARTIC ACID, Asparaginase, CHLORIDE ION | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UPC
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K158M) | | Descriptor: | Asparaginase, CHLORIDE ION, GLYCEROL | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-22 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP3
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21F) | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, Asparaginase, ... | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP9
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K19Q) | | Descriptor: | 1,2-ETHANEDIOL, Asparaginase, CHLORIDE ION, ... | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
