5ODV
 
 | | Structure of Watermelon mosaic virus potyvirus. | | Descriptor: | RNA (5'-R(P*UP*UP*UP*UP*U)-3'), coat protein | | Authors: | Zamora, M, Mendez-Lopez, E, Agirrezabala, X, Cuesta, R, Lavin, J.L, Sanchez-Pina, M.A, Aranda, M, Valle, M. | | Deposit date: | 2017-07-06 | | Release date: | 2017-09-27 | | Last modified: | 2017-10-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Potyvirus virion structure shows conserved protein fold and RNA binding site in ssRNA viruses.
Sci Adv, 3, 2017
|
|
5FN1
 
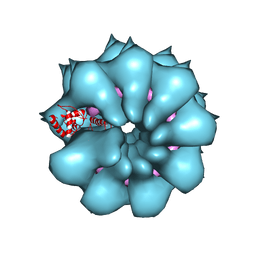 | | Electron cryo-microscopy of filamentous flexible virus PepMV (Pepino Mosaic Virus) | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP)-3', COAT PROTEIN | | Authors: | Agirrezabala, X, Mendez-Lopez, E, Lasso, G, Sanchez-Pina, M.A, Aranda, M.A, Valle, M. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-30 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The near-atomic cryoEM structure of a flexible filamentous plant virus shows homology of its coat protein with nucleoproteins of animal viruses.
Elife, 4, 2015
|
|
5ME6
 
 | | Crystal Structure of eiF4E from C. melo bound to a CAP analog | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic transcription initiation factor 4E | | Authors: | Querol-Audi, J, Silva, C, Miras, M, Aranda-Regules, M, Verdaguer, N. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of eIF4E in Complex with an eIF4G Peptide Supports a Universal Bipartite Binding Mode for Protein Translation.
Plant Physiol., 174, 2017
|
|
5ME7
 
 | | Crystal Structure of eiF4E from C. melo | | Descriptor: | Eukaryotic transcription initiation factor 4E, GLYCEROL | | Authors: | Querol-Audi, J, Silva, C, Miras, M, Aranda-Regules, M, Verdaguer, N. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of eIF4E in Complex with an eIF4G Peptide Supports a Universal Bipartite Binding Mode for Protein Translation.
Plant Physiol., 174, 2017
|
|
5ME5
 
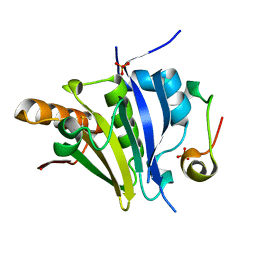 | | Crystal Structure of eiF4E from C. melo bound to a eIF4G peptide | | Descriptor: | Eukaryotic transcription initiation factor 4E, SULFATE ION, eIF4G | | Authors: | Querol-Audi, J, Silva, C, Miras, M, Truniger, V, Aranda-Regules, M, Verdaguer, N. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of eIF4E in Complex with an eIF4G Peptide Supports a Universal Bipartite Binding Mode for Protein Translation.
Plant Physiol., 174, 2017
|
|
5LAF
 
 | |
5LA5
 
 | |
5LA8
 
 | |
5L9J
 
 | |
5LAG
 
 | |
5LAN
 
 | |
