1L17
 
 | |
1L33
 
 | |
1L18
 
 | |
1L24
 
 | |
1L34
 
 | |
1L35
 
 | |
1L19
 
 | |
1L22
 
 | |
1L23
 
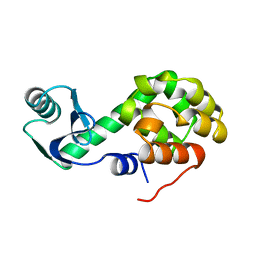 | |
1L20
 
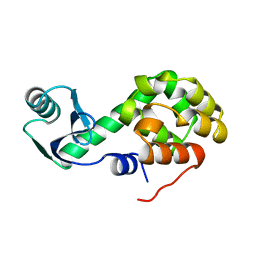 | |
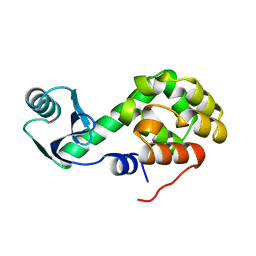
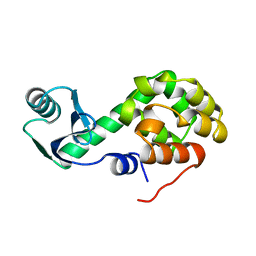
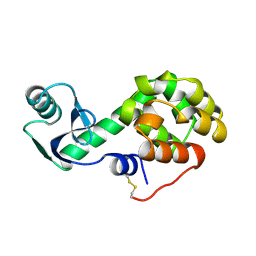
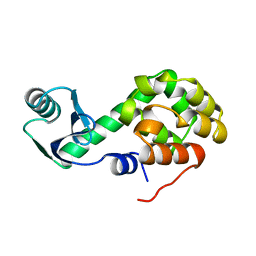
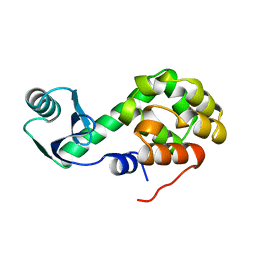
2KLN
 
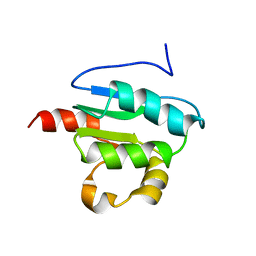 | | Solution Structure of STAS domain of RV1739c from M. tuberculosis | | 分子名称: | PROBABLE SULPHATE-TRANSPORT TRANSMEMBRANE PROTEIN, COG0659 | | 著者 | Sharma, A.K, Ye, L, Zolotarev, A.S, Alper, S.L, Rigby, A.C. | | 登録日 | 2009-07-06 | | 公開日 | 2010-12-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Guanine Nucleotide-binding STAS Domain of SLC26-related SulP Protein Rv1739c from Mycobacterium tuberculosis.
J.Biol.Chem., 286, 2011
|
|
4JLP
 
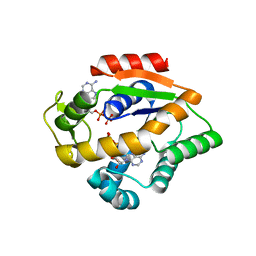 | |
4JLO
 
 | |
4JL6
 
 | |
4JL5
 
 | |
8TIM
 
 | |
4JLD
 
 | |
4JL8
 
 | |
4JLA
 
 | |
4JKY
 
 | |
4JLB
 
 | |
4N8N
 
 | |
3SBO
 
 | | Structure of E.coli GDH from native source | | 分子名称: | CHLORIDE ION, NADP-specific glutamate dehydrogenase | | 著者 | Gee, C.L, Zubieta, C, Echols, N, Totir, M. | | 登録日 | 2011-06-06 | | 公開日 | 2012-03-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.204 Å) | | 主引用文献 | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
3SR0
 
 | |
1CE0
 
 | |
