1ELO
 
 | | ELONGATION FACTOR G WITHOUT NUCLEOTIDE | | 分子名称: | ELONGATION FACTOR G | | 著者 | Aevarsson, A, Brazhnikov, E, Garber, M, Zheltonosova, J, Chirgadze, Yu, Al-Karadaghi, S, Svensson, L.A, Liljas, A. | | 登録日 | 1996-03-13 | | 公開日 | 1996-08-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus.
EMBO J., 13, 1994
|
|
1DTW
 
 | | HUMAN BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE | | 分子名称: | BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE ALPHA SUBUNIT, BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE BETA SUBUNIT, MAGNESIUM ION, ... | | 著者 | AEvarsson, A, Chuang, J.L, Wynn, R.M, Turley, S, Chuang, D.T, Hol, W.G.J. | | 登録日 | 2000-01-13 | | 公開日 | 2000-03-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of human branched-chain alpha-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease.
Structure Fold.Des., 8, 2000
|
|
1QS0
 
 | | Crystal Structure of Pseudomonas Putida 2-oxoisovalerate Dehydrogenase (Branched-Chain Alpha-Keto Acid Dehydrogenase, E1B) | | 分子名称: | 2-OXO-4-METHYLPENTANOIC ACID, 2-OXOISOVALERATE DEHYDROGENASE ALPHA-SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA-SUBUNIT, ... | | 著者 | Aevarsson, A, Seger, K, Turley, S, Sokatch, J.R, Hol, W.G.J. | | 登録日 | 1999-06-24 | | 公開日 | 1999-08-18 | | 最終更新日 | 2021-08-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of 2-oxoisovalerate and dehydrogenase and the architecture of 2-oxo acid dehydrogenase multienzyme complexes.
Nat.Struct.Biol., 6, 1999
|
|
1B5S
 
 | | DIHYDROLIPOYL TRANSACETYLASE (E.C.2.3.1.12) CATALYTIC DOMAIN (RESIDUES 184-425) FROM BACILLUS STEAROTHERMOPHILUS | | 分子名称: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE | | 著者 | Izard, T, Aevarsson, A, Allen, M.D, Westphal, A.H, Perham, R.N, De Kok, A, Hol, W.G. | | 登録日 | 1999-01-10 | | 公開日 | 1999-02-16 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (4.4 Å) | | 主引用文献 | Principles of quasi-equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1DAR
 
 | | ELONGATION FACTOR G IN COMPLEX WITH GDP | | 分子名称: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE | | 著者 | Al-Karadaghi, S, Aevarsson, A, Garber, M, Zheltonosova, J, Liljas, A. | | 登録日 | 1996-02-15 | | 公開日 | 1996-07-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The structure of elongation factor G in complex with GDP: conformational flexibility and nucleotide exchange.
Structure, 4, 1996
|
|
7BGS
 
 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | 分子名称: | Holliday junction resolvase, SULFATE ION | | 著者 | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | 登録日 | 2021-01-08 | | 公開日 | 2022-01-19 | | 最終更新日 | 2022-02-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7BNX
 
 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | 分子名称: | Holliday junction resolvase, SULFATE ION | | 著者 | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | 登録日 | 2021-01-22 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.551 Å) | | 主引用文献 | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R0T
 
 | | Crystal structure of exonuclease ExnV1 | | 分子名称: | CHLORIDE ION, Exonuclease ExnV1, MAGNESIUM ION, ... | | 著者 | Welin, M, Svensson, A, Hakansson, M, Al-Karadaghi, S, Jasilionis, A, Linares-Pasten, J.A, Wang, L, Nordberg Karlsson, E, Ahlqvist, J. | | 登録日 | 2022-02-02 | | 公開日 | 2022-11-02 | | 最終更新日 | 2022-11-23 | | 実験手法 | X-RAY DIFFRACTION (2.194 Å) | | 主引用文献 | Crystal structure of DNA polymerase I from Thermus phage G20c.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R0K
 
 | | Crystal structure of Polymerase I from phage G20c | | 分子名称: | DNA polymerase I | | 著者 | Welin, M, Svensson, A, Hakansson, M, Al-Karadaghi, S, Linares-Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Ahlqvist, J. | | 登録日 | 2022-02-02 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.972 Å) | | 主引用文献 | Crystal structure of DNA polymerase I from Thermus phage G20c.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1FNM
 
 | | STRUCTURE OF THERMUS THERMOPHILUS EF-G H573A | | 分子名称: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | 著者 | Laurberg, M, Kristensen, O, Martemyanov, K, Gudkov, A.T, Nagaev, I, Hughes, D, Liljas, A. | | 登録日 | 2000-08-22 | | 公開日 | 2000-11-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site.
J.Mol.Biol., 303, 2000
|
|
6I5O
 
 | |
6I56
 
 | | Crystal structure of PBSX exported protein XepA | | 分子名称: | GLYCEROL, Phage-like element PBSX protein XepA | | 著者 | Hakansson, M, Svensson, L.A, Welin, M, Al-Karadaghi, S. | | 登録日 | 2018-11-13 | | 公開日 | 2019-11-20 | | 最終更新日 | 2020-11-18 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Crystal structures of the Bacillus subtilis prophage lytic cassette proteins XepA and YomS.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6IA5
 
 | |
487D
 
 | |
7OB6
 
 | |
7OB7
 
 | |
2BM1
 
 | | Ribosomal elongation factor G (EF-G) Fusidic acid resistant mutant G16V | | 分子名称: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | 著者 | Hansson, S, Singh, R, Gudkov, A.T, Liljas, A, Logan, D.T. | | 登録日 | 2005-03-09 | | 公開日 | 2005-05-04 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Insights Into Fusidic Acid Resistance and Sensitivity in EF-G
J.Mol.Biol., 348, 2005
|
|
2BV3
 
 | | Crystal structure of a mutant elongation factor G trapped with a GTP analogue | | 分子名称: | ELONGATION FACTOR G, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Hansson, S, Singh, R, Gudkov, A.T, Liljas, A, Logan, D.T. | | 登録日 | 2005-06-22 | | 公開日 | 2005-08-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of a Mutant Elongation Factor G Trapped with a GTP Analogue.
FEBS Lett., 579, 2005
|
|
2BM0
 
 | | Ribosomal elongation factor G (EF-G) Fusidic acid resistant mutant T84A | | 分子名称: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | 著者 | Hansson, S, Singh, R, Gudkov, A.T, Liljas, A, Logan, D.T. | | 登録日 | 2005-03-09 | | 公開日 | 2005-05-04 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Insights Into Fusidic Acid Resistance and Sensitivity in EF-G
J.Mol.Biol., 348, 2005
|
|
7PJO
 
 | |
2EFG
 
 | | TRANSLATIONAL ELONGATION FACTOR G COMPLEXED WITH GDP | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, PROTEIN (ELONGATION FACTOR G DOMAIN 3), PROTEIN (ELONGATION FACTOR G) | | 著者 | Czworkowski, J, Wang, J, Steitz, T.A, Moore, P.B. | | 登録日 | 1998-09-23 | | 公開日 | 1999-09-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution.
EMBO J., 13, 1994
|
|
1JQM
 
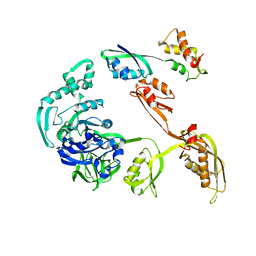 | | Fitting of L11 protein and elongation factor G (EF-G) in the cryo-em map of e. coli 70S ribosome bound with EF-G, GDP and fusidic acid | | 分子名称: | 50S Ribosomal protein L11, Elongation Factor G | | 著者 | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | 登録日 | 2001-08-07 | | 公開日 | 2001-09-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (18 Å) | | 主引用文献 | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
1JQS
 
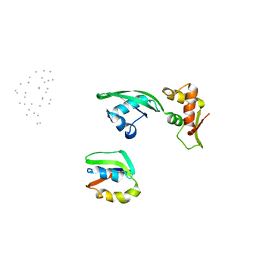 | | Fitting of L11 protein and elongation factor G (domain G' and V) in the cryo-em map of E. coli 70S ribosome bound with EF-G and GMPPCP, a nonhydrolysable GTP analog | | 分子名称: | 50S Ribosomal protein L11, Elongation Factor G | | 著者 | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | 登録日 | 2001-08-07 | | 公開日 | 2001-09-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (18 Å) | | 主引用文献 | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
2BCW
 
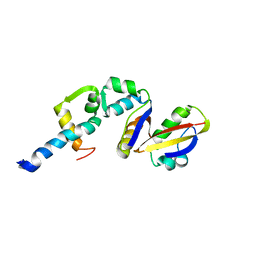 | | Coordinates of the N-terminal domain of ribosomal protein L11,C-terminal domain of ribosomal protein L7/L12 and a portion of the G' domain of elongation factor G, as fitted into cryo-em map of an Escherichia coli 70S*EF-G*GDP*fusidic acid complex | | 分子名称: | 50S ribosomal protein L11, 50S ribosomal protein L7/L12, Elongation factor G | | 著者 | Datta, P.P, Sharma, M.R, Qi, L, Frank, J, Agrawal, R.K. | | 登録日 | 2005-10-19 | | 公開日 | 2005-12-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (11.2 Å) | | 主引用文献 | Interaction of the G' Domain of Elongation Factor G and the C-Terminal Domain of Ribosomal Protein L7/L12 during Translocation as Revealed by Cryo-EM.
Mol.Cell, 20, 2005
|
|
1OLS
 
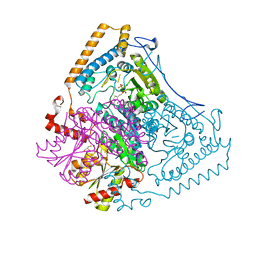 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | 分子名称: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | 著者 | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | 登録日 | 2003-08-12 | | 公開日 | 2003-08-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
