9EPY
 
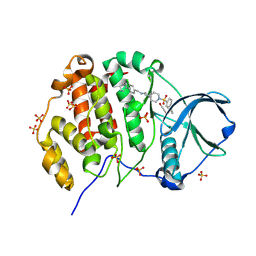 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3330 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3330
To Be Published
|
|
9EPV
 
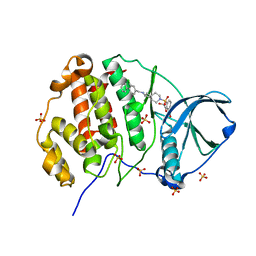 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC333 | | Descriptor: | 1,2-ETHANEDIOL, 5-[[2-[(3-chloranyl-4-phenyl-phenyl)methylamino]-7-azaspiro[3.5]nonan-7-yl]sulfonyl]-1,3-dimethyl-benzimidazol-2-one, Casein kinase II subunit alpha, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC333
To Be Published
|
|
1EWE
 
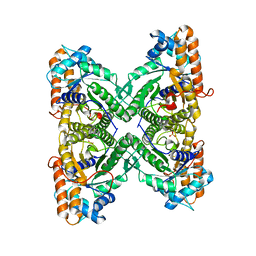 | | Fructose 1,6-Bisphosphate Aldolase from Rabbit Muscle | | Descriptor: | FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE, SULFATE ION | | Authors: | Maurady, A, Sygusch, J. | | Deposit date: | 2000-04-25 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A conserved glutamate residue exhibits multifunctional catalytic roles in
D-fructose-1,6-bisphosphate aldolases.
J.Biol.Chem., 277, 2002
|
|
1EP0
 
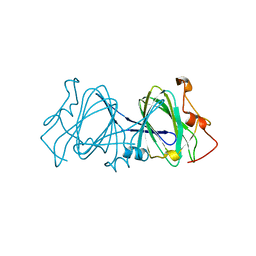 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-24 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
1EPZ
 
 | | CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOASE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM WITH BOUND LIGAND. | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C, Edwards, A.M. | | Deposit date: | 2000-03-30 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
4U2S
 
 | |
4U6H
 
 | | Vaccinia L1/M12B9-Fab complex | | Descriptor: | Heavy chain of murine anti-vaccinia L1 IgG2a antibody M12B9, Light chain of murine anti-vaccinia L1 IgG2a antibody M12B9, Protein L1 | | Authors: | Matho, M.H, Schlossman, A, Zajonc, D.M. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Potent neutralization of vaccinia virus by divergent murine antibodies targeting a common site of vulnerability in l1 protein.
J.Virol., 88, 2014
|
|
4TW1
 
 | | Crystal structure of the octameric pore complex of the Staphylococcus aureus Bi-component Toxin LukGH | | Descriptor: | Possible leukocidin subunit | | Authors: | Logan, D.T, Hakansson, M, Saline, M, Kimbung, R, Badarau, A, Rouha, H, Nagy, E. | | Deposit date: | 2014-06-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Function Analysis of Heterodimer Formation, Oligomerization, and Receptor Binding of the Staphylococcus aureus Bi-component Toxin LukGH.
J.Biol.Chem., 290, 2015
|
|
4U2T
 
 | |
4U2L
 
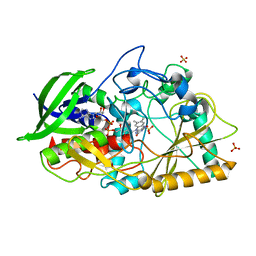 | | Dithionite reduced cholesterol in complex with sulfite | | Descriptor: | (S)-10-((2S,3S,4R)-5-((S)-((S)-(((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHOXY)(HYDROXY)PHOSPHORYLOXY)(HYDROXY)PHOSPHORYLOXY)-2,3,4-TRIHYDROXYPENTYL)-7,8-DIMETHYL-2,4-DIOXO-2,3,4,4A-TETRAHYDROBENZO[G]PTERIDINE-5(10H)-SULFONIC ACID, Cholesterol oxidase, SULFATE ION | | Authors: | Golden, E.A, Vrielink, A. | | Deposit date: | 2014-07-17 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.337 Å) | | Cite: | High-resolution structures of cholesterol oxidase in the reduced state provide insights into redox stabilization.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4UOW
 
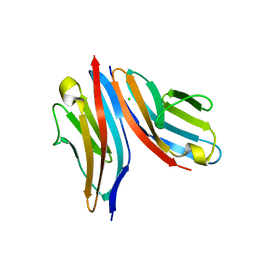 | | Crystal structure of the titin M10-Obscurin Ig domain 1 complex | | Descriptor: | CHLORIDE ION, Obscurin, SODIUM ION, ... | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Human Titin:Obscurin Complex Reveals a Conserved Yet Specific Muscle M-Band Zipper Module.
J.Mol.Biol., 427, 2015
|
|
4UOO
 
 | |
4UOP
 
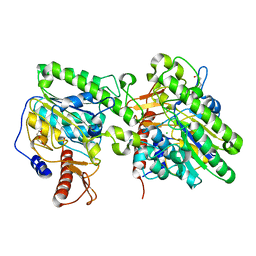 | | Crystal structure of the lipoteichoic acid synthase LtaP from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, LIPOTEICHOIC ACID PRIMASE, MAGNESIUM ION, ... | | Authors: | Campeotto, I, Freemont, P, Grundling, A. | | Deposit date: | 2014-06-06 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and mechanistic insight into the Listeria monocytogenes two-enzyme lipoteichoic acid synthesis system.
J. Biol. Chem., 289, 2014
|
|
4UOR
 
 | | Structure of lipoteichoic acid synthase LtaS from Listeria monocytogenes in complex with glycerol phosphate | | Descriptor: | (2R)-2,3-dihydroxypropyl phosphate, LIPOTEICHOIC ACID SYNTHASE, MAGNESIUM ION | | Authors: | Campeotto, I, Freemont, P, Grundling, A. | | Deposit date: | 2014-06-09 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural and mechanistic insight into the Listeria monocytogenes two-enzyme lipoteichoic acid synthesis system.
J. Biol. Chem., 289, 2014
|
|
4W7O
 
 | |
4W7L
 
 | |
4W63
 
 | | TORPEDO CALIFORNICA ACETYLCHOLINESTERASE IN COMPLEX WITH A TACRINE-BENZOFURAN HYBRID INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Pesaresi, A, Samez, S, Lamba, D. | | Deposit date: | 2014-08-20 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel Tacrine-Benzofuran Hybrids as Potent Multitarget-Directed Ligands for the Treatment of Alzheimer's Disease: Design, Synthesis, Biological Evaluation, and X-ray Crystallography.
J.Med.Chem., 59, 2016
|
|
4V29
 
 | |
4W7N
 
 | |
1DFK
 
 | |
1E3Z
 
 | | Acarbose complex of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.93A | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1E43
 
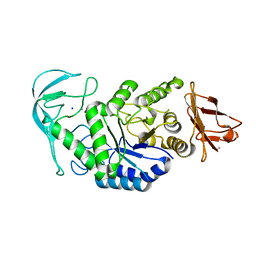 | | Native structure of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.7A | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1E40
 
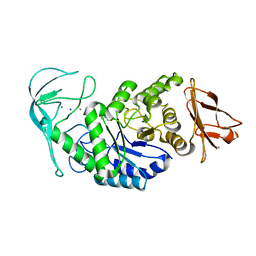 | | Tris/maltotriose complex of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 2.2A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-AMYLASE, CALCIUM ION, ... | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1DAU
 
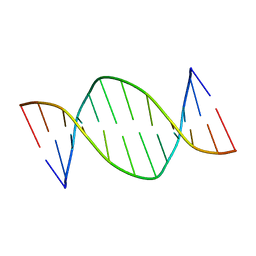 | | Analog of dickerson-drew DNA dodecamer with 6'-alpha-methyl carbocyclic thymidines, NMR, minimized average structure | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(T32)P*(T32)P*CP*GP*CP*G)-3') | | Authors: | Denisov, A, Bekiroglu, S, Maltseva, T, Sandstrom, A, Altmann, K.-H, Egli, M, Chattopadhyaya, J. | | Deposit date: | 1998-01-21 | | Release date: | 1998-05-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution conformation of a carbocyclic analog of the Dickerson-Drew dodecamer: comparison with its own X-ray structure and that of the NMR structure of the native counterpart.
J.Biomol.Struct.Dyn., 16, 1998
|
|
1DNP
 
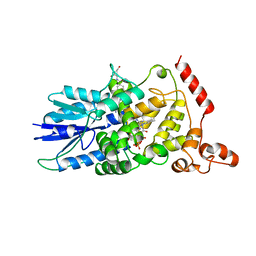 | | STRUCTURE OF DEOXYRIBODIPYRIMIDINE PHOTOLYASE | | Descriptor: | 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA PHOTOLYASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Park, H.-W, Sancar, A, Deisenhofer, J. | | Deposit date: | 1995-07-03 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of DNA photolyase from Escherichia coli.
Science, 268, 1995
|
|
