4EZR
 
 | |
4EZW
 
 | |
4EZO
 
 | |
4EZZ
 
 | |
4F00
 
 | |
4EZQ
 
 | |
4EZU
 
 | |
4EZS
 
 | |
4EZY
 
 | |
4EZN
 
 | |
4EZX
 
 | |
4EZV
 
 | |
8C65
 
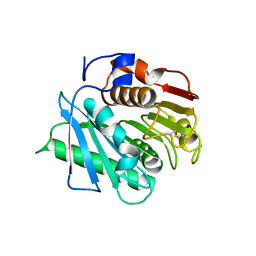 | |
8AYV
 
 | |
8AIT
 
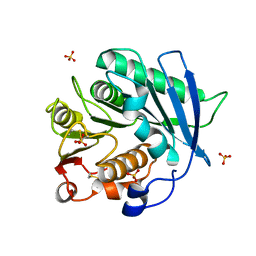 | | Crystal structure of cutinase PbauzCut from Pseudomonas bauzanensis | | Descriptor: | Cutinase, SULFATE ION | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
8AIS
 
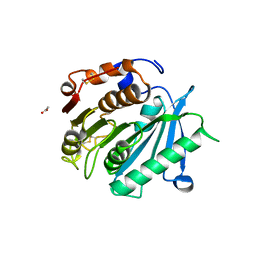 | | Crystal structure of cutinase PsCut from Pseudomonas saudimassiliensis | | Descriptor: | ACETATE ION, Lipase 1 | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
8AIR
 
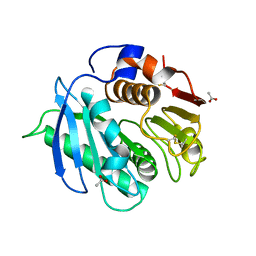 | | Crystal structure of cutinase RgCutII from Rhizobacter gummiphilus | | Descriptor: | ACETATE ION, RgCutII | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
6EUS
 
 | |
4R3U
 
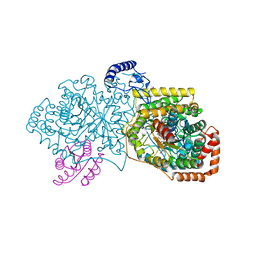 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Mutase | | Descriptor: | 2-hydroxyisobutyryl-CoA mutase large subunit, 2-hydroxyisobutyryl-CoA mutase small subunit, 3-HYDROXYBUTANOYL-COENZYME A, ... | | Authors: | Zahn, M, Kurteva-Yaneva, N, Rohwerder, T, Straeter, N. | | Deposit date: | 2014-08-18 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the stereospecificity of bacterial B12-dependent 2-hydroxyisobutyryl-CoA mutase.
J.Biol.Chem., 290, 2015
|
|
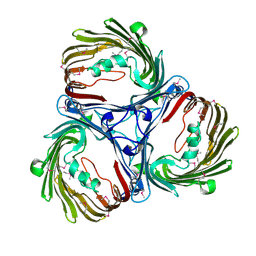
5MDP
 
 | |
5MDS
 
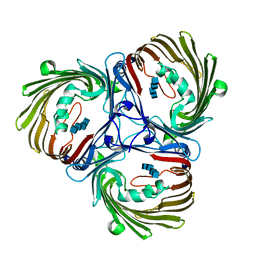 | |
5MDR
 
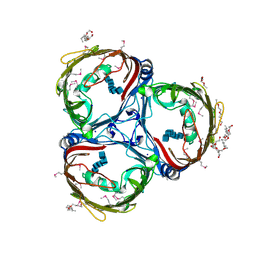 | | Crystal structure of in vitro folded Chitoporin VhChip from Vibrio harveyi in complex with chitohexaose | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitoporin, ... | | Authors: | Zahn, M, van den Berg, B. | | Deposit date: | 2016-11-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for chitin acquisition by marine Vibrio species.
Nat Commun, 9, 2018
|
|
5MDO
 
 | |
5MDQ
 
 | |
3QNJ
 
 | |
