3A86
 
 | |
2ZY6
 
 | | Crystal structure of a truncated tRNA, TPHE39A | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Tanaka, I, Yao, M, Tanaka, Y, Kitago, Y, Ymagata, S. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deduced RNA binding mechanism of ThiI based on structural and binding analyses of a minimal RNA ligand
Rna, 15, 2009
|
|
3A7Y
 
 | |
3A8A
 
 | |
3A89
 
 | |
3A84
 
 | |
3ANZ
 
 | | Crystal Structure of alpha-hemolysin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Alpha-hemolysin | | Authors: | Yamashita, K, Kawauchi, H, Tanaka, Y, Yao, M, Tanaka, I. | | Deposit date: | 2010-09-16 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | 2-Methyl-2,4-pentanediol induces spontaneous assembly of staphylococcal alpha-hemolysin into heptameric pore structure
Protein Sci., 20, 2011
|
|
3A7Z
 
 | |
3A8B
 
 | |
3ATK
 
 | | Crystal structure of trypsin complexed with cycloheptanamine | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Tanaka, I. | | Deposit date: | 2011-01-05 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3ATI
 
 | | Crystal structure of trypsin complexed with (3-methoxyphenyl)methanamine | | Descriptor: | 1-(3-methoxyphenyl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Tanaka, I. | | Deposit date: | 2011-01-05 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3ATM
 
 | | Crystal structure of trypsin complexed with 2-(1H-indol-3-yl)ethanamine | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Tanaka, I. | | Deposit date: | 2011-01-05 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3ATL
 
 | | Crystal structure of trypsin complexed with benzamidine | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Tanaka, I. | | Deposit date: | 2011-01-05 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3B0V
 
 | | tRNA-dihydrouridine synthase from Thermus thermophilus in complex with tRNA | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA, tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-14 | | Release date: | 2011-12-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3B0U
 
 | | tRNA-dihydrouridine synthase from Thermus thermophilus in complex with tRNA fragment | | Descriptor: | FLAVIN MONONUCLEOTIDE, RNA (5'-R(*GP*GP*(H2U)P*A)-3'), tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-14 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3B0P
 
 | | tRNA-dihydrouridine synthase from Thermus thermophilus | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-12 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AZV
 
 | | Crystal structure of the receptor binding domain | | Descriptor: | D/C mosaic neurotoxin, SULFATE ION | | Authors: | Nuemket, N, Tanaka, Y, Tsukamoto, K, Tsuji, T, Nakamura, K, Kozaki, S, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-02 | | Release date: | 2011-12-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and mutational analyses of the receptor binding domain of botulinum D/C mosaic neurotoxin: insight into the ganglioside binding mechanism
Biochem.Biophys.Res.Commun., 411, 2011
|
|
3AZW
 
 | | Crystal structure of the receptor binding domain | | Descriptor: | D/C mosaic neurotoxin, SULFATE ION | | Authors: | Nuemket, N, Tanaka, Y, Tsukamoto, K, Tsuji, T, Nakamura, K, Kozaki, S, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-02 | | Release date: | 2011-12-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural and mutational analyses of the receptor binding domain of botulinum D/C mosaic neurotoxin: insight into the ganglioside binding mechanism
Biochem.Biophys.Res.Commun., 411, 2011
|
|
6LIU
 
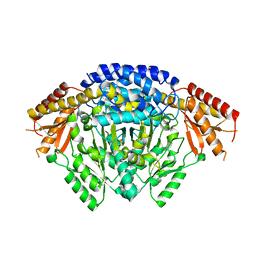 | | Crystal structure of apo Tyrosine decarboxylase | | Descriptor: | Tyrosine/DOPA decarboxylase 2 | | Authors: | Yu, J, Wang, H, Yao, M. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures clarify cofactor binding of plant tyrosine decarboxylase.
Biochem.Biophys.Res.Commun., 2019
|
|
6LIV
 
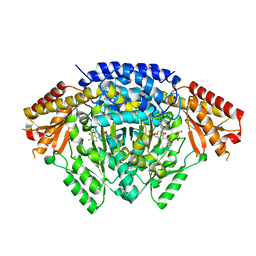 | |
3WSU
 
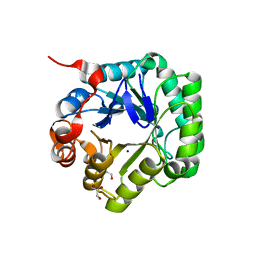 | | Crystal structure of beta-mannanase from Streptomyces thermolilacinus | | Descriptor: | Beta-mannanase, GLYCEROL, SODIUM ION | | Authors: | Kumagai, Y, Yamashita, K, Okuyama, M, Hatanaka, T, Yao, M, Kimura, A. | | Deposit date: | 2014-03-26 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition
Febs J., 282, 2015
|
|
8H25
 
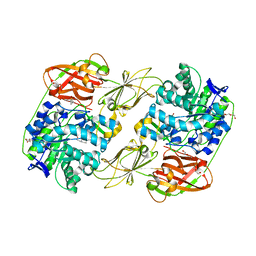 | | Lacticaseibacillus casei GH35 beta-galactosidase LBCZ_0230 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-galactosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saburi, W, Ota, T, Kato, K, Tagami, T, Yamashita, K, Yao, M, Mori, H. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Function and Structure of Lacticaseibacillus casei GH35 beta-Galactosidase LBCZ_0230 with High Hydrolytic Activity to Lacto- N -biose I and Galacto- N -biose.
J Appl Glycosci (1999), 70, 2023
|
|
5X32
 
 | | Crystal structure of D-mannose isomerase | | Descriptor: | N-acylglucosamine 2-epimerase, PHOSPHATE ION | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Biochemical and structural characterization of Marinomonas mediterranead-mannose isomerase Marme_2490 phylogenetically distant from known enzymes
Biochimie, 144, 2018
|
|
5XW7
 
 | |
8HRH
 
 | | SN-131/1B2 anti-MUC1 antibody with a glycopeptide | | Descriptor: | 1-ACETYL-L-PROLINE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALANINE, ... | | Authors: | Wakui, H, Horidome, C, Yao, M, Ose, T, Nishimura, S.-I. | | Deposit date: | 2022-12-15 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural and molecular insight into antibody recognition of dynamic neoepitopes in membrane tethered MUC1 of pancreatic cancer cells and secreted exosomes.
Rsc Chem Biol, 4, 2023
|
|
