2ZVU
 
 | | Crystal structure of rat heme oxygenase-1 in complex with ferrous verdoheme | | Descriptor: | 5-OXA-PROTOPORPHYRIN IX CONTAINING FE, FORMIC ACID, Heme oxygenase 1 | | Authors: | Sato, H, Sugishima, M, Fukuyama, K, Noguchi, M. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of rat haem oxygenase-1 in complex with ferrous verdohaem: presence of a hydrogen-bond network on the distal side
Biochem.J., 419, 2009
|
|
2D1E
 
 | | Crystal structure of PcyA-biliverdin complex | | Descriptor: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase, SODIUM ION | | Authors: | Hagiwara, Y, Sugishima, M, Takahashi, Y, Fukuyama, K. | | Deposit date: | 2005-08-17 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin IXalpha, a key enzyme in the biosynthesis of phycocyanobilin
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DKE
 
 | | Crystal structure of substrate-free form of PcyA | | Descriptor: | CHLORIDE ION, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Hagiwara, Y, Sugishima, M, Takahashi, Y, Fukuyama, K. | | Deposit date: | 2006-04-10 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Induced-fitting and electrostatic potential change of PcyA upon substrate binding demonstrated by the crystal structure of the substrate-free form
Febs Lett., 580, 2006
|
|
1V47
 
 | | Crystal structure of ATP sulfurylase from Thermus thermophillus HB8 in complex with APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, ATP sulfurylase, CHLORIDE ION, ... | | Authors: | Taguchi, Y, Sugishima, M, Fukuyama, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of a novel zinc-binding ATP sulfurylase from Thermus thermophilus HB8
Biochemistry, 43, 2004
|
|
7YL8
 
 | |
5B3T
 
 | |
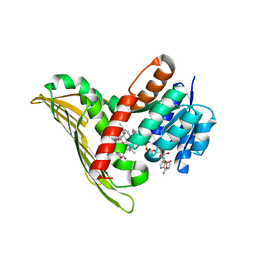
5B3V
 
 | |
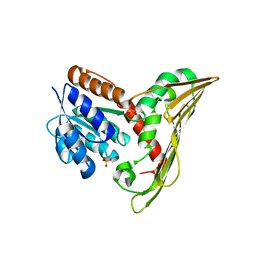
5B3U
 
 | |
7YKB
 
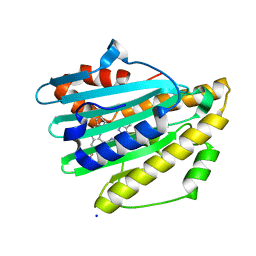 | | Neutron Structure of PcyA D105N Mutant Complexed with Biliverdin at Room Temperature | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Phycocyanobilin:ferredoxin oxidoreductase, SODIUM ION | | Authors: | Unno, M, Nanasawa, R. | | Deposit date: | 2022-07-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.38 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography and quantum chemical analysis of bilin reductase PcyA mutants reveal substrate and catalytic residue protonation states.
J.Biol.Chem., 299, 2022
|
|
7YK9
 
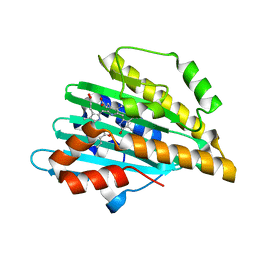 | | Neutron Structure of PcyA I86D Mutant Complexed with Biliverdin at Room Temperature | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Unno, M, Igarashi, K. | | Deposit date: | 2022-07-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography and quantum chemical analysis of bilin reductase PcyA mutants reveal substrate and catalytic residue protonation states.
J.Biol.Chem., 299, 2022
|
|
