1GKM
 
 | |
1GKY
 
 | |
1GES
 
 | |
1GEU
 
 | | ANATOMY OF AN ENGINEERED NAD-BINDING SITE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mittl, P.R.E, Schulz, G.E. | | Deposit date: | 1994-01-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anatomy of an engineered NAD-binding site.
Protein Sci., 3, 1994
|
|
1GET
 
 | | ANATOMY OF AN ENGINEERED NAD-BINDING SITE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mittl, P.R.E, Schulz, G.E. | | Deposit date: | 1994-01-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anatomy of an engineered NAD-binding site.
Protein Sci., 3, 1994
|
|
1GK3
 
 | |
1GK2
 
 | |
1GQC
 
 | |
1GQ9
 
 | |
2BHS
 
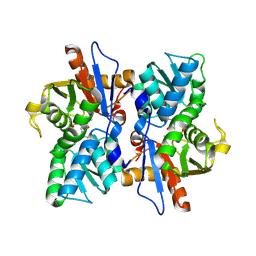 | | Crystal Structure of Cysteine Synthase B | | Descriptor: | CYSTEINE SYNTHASE B, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Claus, M.T, Zocher, G.E, Maier, T.H.P, Schulz, G.E. | | Deposit date: | 2005-01-18 | | Release date: | 2005-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structure of the O-Acetylserine Sulfhydrylase Isoenzyme Cysm from Escherichia Coli
Biochemistry, 44, 2005
|
|
1PAX
 
 | |
2BHT
 
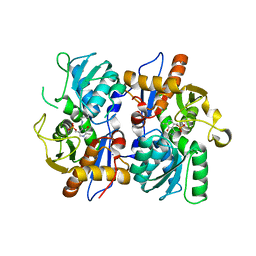 | | Crystal structure of O-acetylserine sulfhydrylase B | | Descriptor: | CYSTEINE SYNTHASE B, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Claus, M.T, Zocher, G.E, Maier, T.H.P, Schulz, G.E. | | Deposit date: | 2005-01-18 | | Release date: | 2005-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the O-Acetylserine Sulfhydrylase Isoenzyme Cysm from Escherichia Coli
Biochemistry, 44, 2005
|
|
1POW
 
 | |
1POX
 
 | |
1PRN
 
 | |
1QJ9
 
 | |
1QJP
 
 | |
1QJ8
 
 | |
1SQC
 
 | |
1UKY
 
 | |
5PRN
 
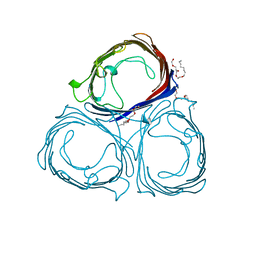 | | E1M, Y96W, S119W MUTANT OF RH. BLASTICA PORIN | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, PORIN | | Authors: | Maveyraud, L, Schmid, B, Schulz, G.E. | | Deposit date: | 1998-06-12 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Porin mutants with new channel properties.
Protein Sci., 7, 1998
|
|
2V2B
 
 | |
2J1P
 
 | | Geranylgeranyl diphosphate synthase from Sinapis alba in complex with GGPP | | Descriptor: | BETA-MERCAPTOETHANOL, GERANYLGERANYL DIPHOSPHATE, GERANYLGERANYL PYROPHOSPHATE SYNTHETASE, ... | | Authors: | Kloer, D.P, Welsch, R, Beyer, P, Schulz, G.E. | | Deposit date: | 2006-08-15 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Reaction Geometry of Geranylgeranyl Diphosphate Synthase from Sinapis Alba.
Biochemistry, 45, 2006
|
|
2J1O
 
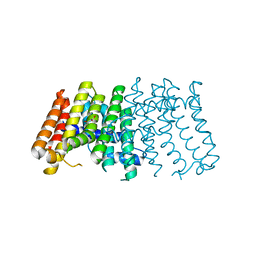 | | Geranylgeranyl diphosphate synthase from Sinapis alba | | Descriptor: | GERANYLGERANYL PYROPHOSPHATE SYNTHETASE, GLYCEROL | | Authors: | Kloer, D.P, Welsch, R, Beyer, P, Schulz, G.E. | | Deposit date: | 2006-08-15 | | Release date: | 2007-01-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Reaction Geometry of Geranylgeranyl Diphosphate Synthase from Sinapis Alba.
Biochemistry, 45, 2006
|
|
2IVF
 
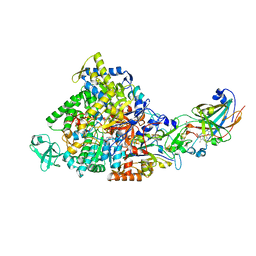 | | Ethylbenzene dehydrogenase from Aromatoleum aromaticum | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ACETATE ION, ... | | Authors: | Kloer, D.P, Hagel, C, Heider, J, Schulz, G.E. | | Deposit date: | 2006-06-13 | | Release date: | 2006-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Ethylbenzene Dehydrogenase from Aromatoleum Aromaticum
Structure, 14, 2006
|
|
