3X2G
 
 | | X-ray structure of PcCel45A N92D apo form at 100K | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | 著者 | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | 登録日 | 2014-12-22 | | 公開日 | 2015-10-07 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2L
 
 | | X-ray structure of PcCel45A apo form at 95K. | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | 著者 | Nakamura, A, Ishida, T, Ohta, K, Tanaka, H, Inaka, K, Samejima, M, Igarashi, K. | | 登録日 | 2014-12-22 | | 公開日 | 2015-10-14 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (0.83 Å) | | 主引用文献 | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2N
 
 | | Proton relay pathway in inverting cellulase | | 分子名称: | Endoglucanase V-like protein, SULFATE ION | | 著者 | Nakamura, A, Ishida, T, Fushinobu, S, Igarashi, K, Samejima, M. | | 登録日 | 2014-12-22 | | 公開日 | 2015-10-14 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
7VC7
 
 | | The structure of beta-xylosidase from Phanerochaete chrysosporium(PcBxl3) | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kojima, K, Sunagawa, N, Igarashi, K. | | 登録日 | 2021-09-01 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Comparison of glycoside hydrolase family 3 beta-xylosidases from basidiomycetes and ascomycetes reveals evolutionarily distinct xylan degradation systems.
J.Biol.Chem., 298, 2022
|
|
7VC6
 
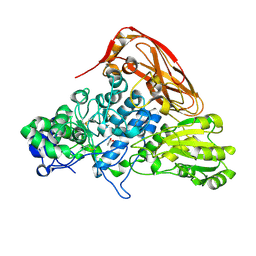 | | The structure of beta-xylosidase from Phanerochaete chrysosporium(PcBxl3) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, xylan 1,4-beta-xylosidase | | 著者 | Kojima, K, Sunagawa, N, Igarashi, K. | | 登録日 | 2021-09-01 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Comparison of glycoside hydrolase family 3 beta-xylosidases from basidiomycetes and ascomycetes reveals evolutionarily distinct xylan degradation systems.
J.Biol.Chem., 298, 2022
|
|
3A64
 
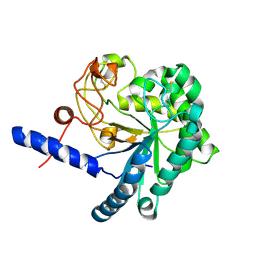 | | Crystal structure of CcCel6C, a glycoside hydrolase family 6 enzyme, from Coprinopsis cinerea | | 分子名称: | Cellobiohydrolase, MAGNESIUM ION | | 著者 | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | 登録日 | 2009-08-21 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
3A9B
 
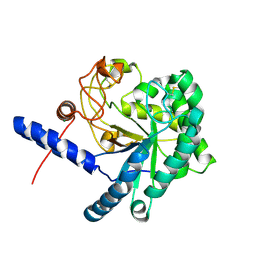 | | CcCel6C, a glycoside hydrolase family 6 enzyme, complexed with cellobiose | | 分子名称: | Cellobiohydrolase, MAGNESIUM ION, beta-D-glucopyranose, ... | | 著者 | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | 登録日 | 2009-10-22 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
3ABX
 
 | | CcCel6C, a glycoside hydrolase family 6 enzyme, complexed with p-nitrophenyl beta-D-cellotrioside | | 分子名称: | 4-nitrophenyl beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranoside, Cellobiohydrolase, MAGNESIUM ION | | 著者 | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | 登録日 | 2009-12-24 | | 公開日 | 2010-01-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
5YSD
 
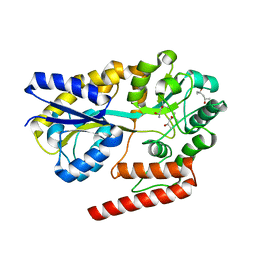 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotriose | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | 著者 | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | 登録日 | 2017-11-14 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSF
 
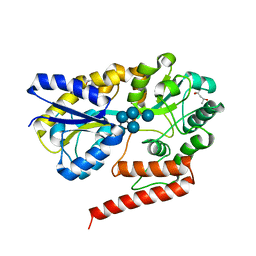 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophoropentaose | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | 著者 | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | 登録日 | 2017-11-14 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSE
 
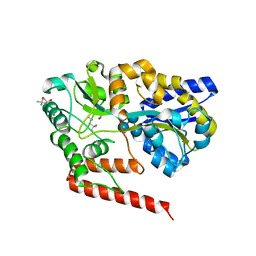 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotetraose | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | 著者 | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | 登録日 | 2017-11-14 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSB
 
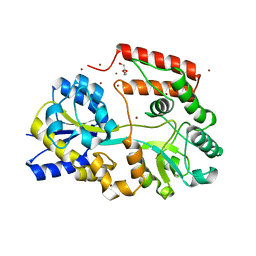 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in ligand-free form | | 分子名称: | DI(HYDROXYETHYL)ETHER, Lin1841 protein, ZINC ION | | 著者 | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | 登録日 | 2017-11-13 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
7BYT
 
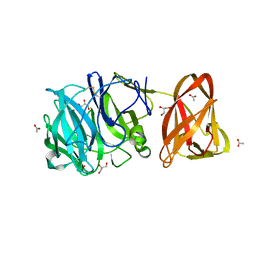 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A with galactose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | 登録日 | 2020-04-24 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
7BYX
 
 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A E208A with beta-1,3-galactotriose | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | 登録日 | 2020-04-24 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
7BYS
 
 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A apo form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CITRIC ACID, ... | | 著者 | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | 登録日 | 2020-04-24 | | 公開日 | 2020-11-04 | | 最終更新日 | 2021-01-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
7BYV
 
 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A E208Q with beta-1,3-galactotriose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Galactan 1,3-beta-galactosidase, ... | | 著者 | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | 登録日 | 2020-04-24 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
4B5Q
 
 | | The lytic polysaccharide monooxygenase GH61D structure from the basidiomycota fungus Phanerochaete chrysosporium | | 分子名称: | COPPER (II) ION, GLYCEROL, GLYCOSIDE HYDROLASE FAMILY 61 PROTEIN D, ... | | 著者 | Wu, M, Beckham, G.T, Larsson, A.M, Ishida, T, Kim, S, Crowley, M.F, Payne, C.M, Horn, S.J, Westereng, B, Stahlberg, J, Eijsink, V.G.H, Sandgren, M. | | 登録日 | 2012-08-07 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structure and Computational Characterization of the Lytic Polysaccharide Monooxygenase Gh61D from the Basidiomycota Fungus Phanerochaete Chrysosporium
J.Biol.Chem., 288, 2013
|
|
5X6S
 
 | | Acetyl xylan esterase from Aspergillus awamori | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylxylan esterase A, ... | | 著者 | Komiya, D, Koseki, T, Fushinobu, S. | | 登録日 | 2017-02-23 | | 公開日 | 2017-08-23 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure and Substrate Specificity Modification of Acetyl Xylan Esterase from Aspergillus luchuensis
Appl. Environ. Microbiol., 83, 2017
|
|
3WMT
 
 | | Crystal structure of feruloyl esterase B from Aspergillus oryzae | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Probable feruloyl esterase B-1 | | 著者 | Suzuki, K, Ishida, T, Igarashi, K, Koseki, T, Fushinobu, S. | | 登録日 | 2013-11-25 | | 公開日 | 2014-08-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of a feruloyl esterase belonging to the tannase family: a disulfide bond near a catalytic triad.
Proteins, 82, 2014
|
|
