3P9C
 
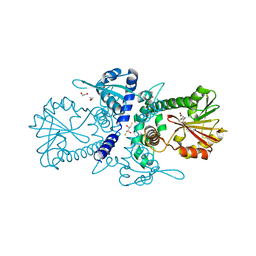 | | Crystal structure of perennial ryegrass LpOMT1 bound to SAH | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Louie, G.V, Noel, J.P, Bowman, M.E. | | Deposit date: | 2010-10-17 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Analyses of a Caffeic Acid O-Methyltransferase from Perennial Ryegrass Reveal the Molecular Basis for Substrate Preference.
Plant Cell, 22, 2010
|
|
3P9I
 
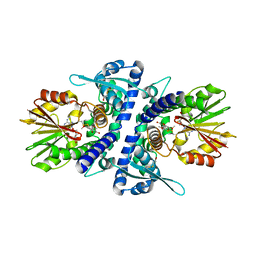 | | Crystal structure of perennial ryegrass LpOMT1 complexed with S-adenosyl-L-homocysteine and sinapaldehyde | | Descriptor: | (2E)-3-(4-hydroxy-3,5-dimethoxyphenyl)prop-2-enal, BETA-MERCAPTOETHANOL, Caffeic acid O-methyltransferase, ... | | Authors: | Louie, G.V, Noel, J.P, Bowman, M.E. | | Deposit date: | 2010-10-17 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Function Analyses of a Caffeic Acid O-Methyltransferase from Perennial Ryegrass Reveal the Molecular Basis for Substrate Preference.
Plant Cell, 22, 2010
|
|
3QTC
 
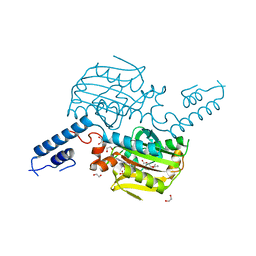 | | Crystal structure of the catalytic domain of MmOmeRS, an O-methyl tyrosyl-tRNA synthetase evolved from Methanosarcina mazei PylRS, complexed with O-methyl tyrosine and AMP-PNP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, O-methyl-L-tyrosine, ... | | Authors: | Dellas, N, Takimoto, J.K, Noel, J.P, Wang, L. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Stereochemical Basis for Engineered Pyrrolysyl-tRNA Synthetase and the Efficient in Vivo Incorporation of Structurally Divergent Non-native Amino Acids.
Acs Chem.Biol., 6, 2011
|
|
3STU
 
 | |
3STT
 
 | |
3STV
 
 | |
3STW
 
 | |
3STY
 
 | |
3STX
 
 | |
1Y6I
 
 | | Synechocystis GUN4 | | Descriptor: | Mg-chelatase cofactor GUN4 | | Authors: | Verdecia, M.A, Larkin, R.M, Ferrer, J.L, Riek, R, Chory, J, Noel, J.P. | | Deposit date: | 2004-12-06 | | Release date: | 2005-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the Mg-chelatase cofactor GUN4 reveals a novel hand-shaped fold for porphyrin binding
Plos Biol., 3, 2005
|
|
1YQD
 
 | | Sinapyl Alcohol Dehydrogenase complexed with NADP+ | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Bomati, E.K, Noel, J.P. | | Deposit date: | 2005-02-01 | | Release date: | 2005-07-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Kinetic Basis for Substrate Selectivity in Populus tremuloides Sinapyl Alcohol Dehydrogenase.
Plant Cell, 17, 2005
|
|
1YQX
 
 | | Sinapyl Alcohol Dehydrogenase at 2.5 Angstrom Resolution | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION, sinapyl alcohol dehydrogenase | | Authors: | Bomati, E.K, Noel, J.P. | | Deposit date: | 2005-02-02 | | Release date: | 2005-07-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Kinetic Basis for Substrate Selectivity in Populus tremuloides Sinapyl Alcohol Dehydrogenase.
Plant Cell, 17, 2005
|
|
1YFO
 
 | |
1ZGA
 
 | | Crystal structure of isoflavanone 4'-O-methyltransferase complexed with (+)-6a-hydroxymaackiain | | Descriptor: | (6AR,12AR)-6H-[1,3]DIOXOLO[5,6][1]BENZOFURO[3,2-C]CHROMENE-3,6A(12AH)-DIOL, Isoflavanone 4'-O-methyltransferase', S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-20 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
1ZB6
 
 | | Co-Crystal Structure of ORF2 an Aromatic Prenyl Transferase from Streptomyces sp. strain cl190 complexed with GSPP and 1,6-dihydroxynaphtalene | | Descriptor: | 1,6-DIHYDROXY NAPHTHALENE, Aromatic prenyltransferase, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Kuzuyama, T, Noel, J.P, Richard, S.B. | | Deposit date: | 2005-04-07 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products.
Nature, 435, 2005
|
|
1ZCN
 
 | | human Pin1 Ng mutant | | Descriptor: | PENTAETHYLENE GLYCOL, PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jager, M, Zhang, Y, Nguyen, H, Dendel, G, Bowman, M.E, Gruebele, M, Noel, J.P, Kelly, J.W. | | Deposit date: | 2005-04-12 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function-folding relationship in a WW domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1ZDW
 
 | | Co-crystal structure of Orf2 an aromatic prenyl transferase from Streptomyces sp. strain CL190 complexed with GSPP and Flaviolin | | Descriptor: | Aromatic prenyltransferase, FLAVIOLIN, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Kuzuyama, T, Noel, J.P, Richard, S.B. | | Deposit date: | 2005-04-15 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products.
Nature, 435, 2005
|
|
1ZG3
 
 | | Crystal structure of the isoflavanone 4'-O-methyltransferase complexed with SAH and 2,7,4'-trihydroxyisoflavanone | | Descriptor: | (2S,3R)-2,7-DIHYDROXY-3-(4-HYDROXYPHENYL)-2,3-DIHYDRO-4H-CHROMEN-4-ONE, S-ADENOSYL-L-HOMOCYSTEINE, isoflavanone 4'-O-methyltransferase | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-20 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
1ZDY
 
 | |
1ZHF
 
 | | Crystal structure of selenomethionine substituted isoflavanone 4'-O-methyltransferase | | Descriptor: | Isoflavanone 4'-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-25 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
1ZCW
 
 | | Co-crystal structure of Orf2 an aromatic prenyl transferase from Streptomyces sp. strain CL190 complexed with GPP | | Descriptor: | Aromatic prenyltransferase, GERANYL DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kuzuyama, T, Noel, J.P, Richard, S.B. | | Deposit date: | 2005-04-13 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products.
Nature, 435, 2005
|
|
1ZGJ
 
 | | Crystal structure of isoflavanone 4'-O-methyltransferase complexed with (+)-pisatin | | Descriptor: | (6AR,12AR)-3-(HYDROXYMETHYL)-6H-[1,3]DIOXOLO[5,6][1]BENZOFURO[3,2-C]CHROMEN-6A(12AH)-OL, Isoflavanone 4'-O-methyltransferase', S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-21 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
1ZGD
 
 | | Chalcone Reductase Complexed With NADP+ at 1.7 Angstrom Resolution | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, chalcone reductase | | Authors: | Bomati, E.K, Austin, M.B, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-21 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural elucidation of chalcone reductase and implications for deoxychalcone biosynthesis
J.Biol.Chem., 280, 2005
|
|
2Q6K
 
 | | SalL with adenosine | | Descriptor: | ADENOSINE, DI(HYDROXYETHYL)ETHER, chlorinase | | Authors: | Pojer, F, Noel, J.P. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and characterization of a marine bacterial SAM-dependent chlorinase
Nat.Chem.Biol., 4, 2008
|
|
2Q6L
 
 | | SalL double mutant Y70T/G131S with CLDA and L-MET | | Descriptor: | 5'-CHLORO-5'-DEOXYADENOSINE, Hypothetical protein, METHIONINE | | Authors: | Pojer, F, Noel, J.P. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Discovery and characterization of a marine bacterial SAM-dependent chlorinase
Nat.Chem.Biol., 4, 2008
|
|
